3V8R
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-amino-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V80
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-O-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-O-prop-2-yn-1-yladenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V7W
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-azido-5'-deoxyadenosine | | Descriptor: | 5'-azido-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.0102 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8M
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with 5'-azido-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8Q
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-5'-deoxyadenosine | | Descriptor: | 5'-amino-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
6V6Q
 
 | | Crystal Structure of Monophosphorylated FGF Receptor 2 isoform IIIb with PTR657 | | Descriptor: | Fibroblast growth factor receptor 2, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lin, C.-C, Wieteska, L, Poncet-Montange, G, Suen, K.M, Arold, S.T, Ahmed, Z, Ladbury, J.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The combined action of the intracellular regions regulates FGFR2 kinase activity
Commun Biol, 6, 2023
|
|
4DY6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 2'-phosphate bis(adenosine)-5'-diphosphate | | Descriptor: | CITRIC ACID, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
3V8P
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a new di-adenosine inhibitor formed in situ | | Descriptor: | 2-[6-azanyl-9-[(2R,3R,4S,5R)-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]sulfanyl-N-[[(2R,3S,4R,5R)-5-(6-azanyl-8-bromanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]ethanamide, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2901 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V7Y
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-N-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-deoxy-5'-(prop-2-yn-1-ylamino)adenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
2Q5F
 
 | | Crystal structure of LMNADK1 from Listeria monocytogenes | | Descriptor: | (2S,3S,4R,5R,2'S,3'S,4'R,5'R)-2,2'-[DITHIOBIS(METHYLENE)]BIS[5-(6-AMINO-9H-PURIN-9-YL)TETRAHYDROFURAN-3,4-DIOL], Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2007-06-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2NTN
 
 | | Crystal structure of MabA-C60V/G139A/S144L | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Poncet-Montange, G, Ducasse-Cabanot, S, Quemard, A, Labesse, G, Cohen-Gonsaud, M. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of dynamics in the MabA active site kills the enzyme activity: practical consequences for drug-design studies
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4TU4
 
 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT4
 
 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT2
 
 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT6
 
 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU6
 
 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
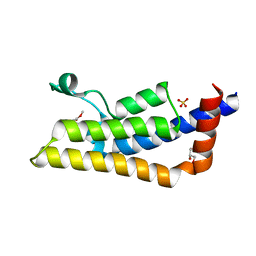 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
2I2F
 
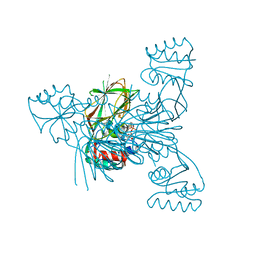 | | Crystal structure of LmNADK1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2I2B
 
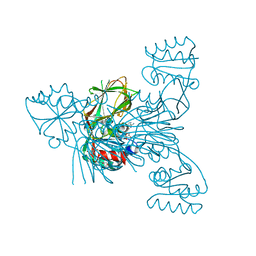 | |
2I2D
 
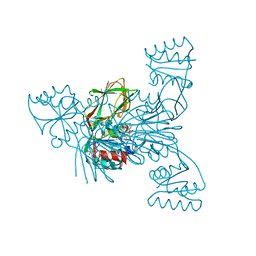 | | Crystal structure of LmNADK1 | | Descriptor: | BIS{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL} DIHYDROGEN DIPHOSPHATE, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2I1W
 
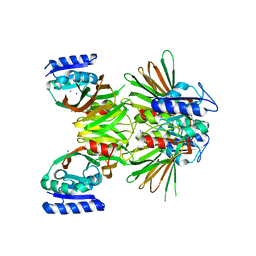 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes | | Descriptor: | IODIDE ION, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2I2C
 
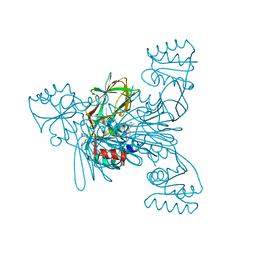 | | Crystal structure of LmNADK1 | | Descriptor: | (2S,3S,4R,5R,2'S,3'S,4'R,5'R)-2,2'-[DITHIOBIS(METHYLENE)]BIS[5-(6-AMINO-9H-PURIN-9-YL)TETRAHYDROFURAN-3,4-DIOL], Probable inorganic polyphosphate/ATP-NAD kinase 1, TETRAETHYLENE GLYCOL | | Authors: | Poncet-Montange, G, Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2I29
 
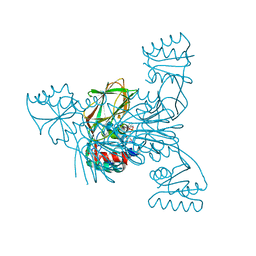 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
