4YIL
 
 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL 3-CYANO-3-(4-FLUOROPHENYL)ACRYLATE IN A NON PRODUCTIVE BINDING MODE | | 分子名称: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH dehydrogenase 1, ... | | 著者 | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | 登録日 | 2015-03-02 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.461 Å) | | 主引用文献 | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
2VZN
 
 | |
5AYL
 
 | | Crystal structure of ERdj5 form II | | 分子名称: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DnaJ homolog subfamily C member 10 | | 著者 | Watanabe, S, Maegawa, K, Inaba, K. | | 登録日 | 2015-08-22 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
2W27
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | 分子名称: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | 著者 | Padavattan, S, AndERSON, W.F, Schirmer, T. | | 登録日 | 2008-10-24 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
5AZZ
 
 | | Crystal structure of seleno-insulin | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Watanabe, S, Okumura, M, Arai, K, Takei, T, Asahina, Y, Hojo, H, Iwaoka, M, Inaba, K. | | 登録日 | 2015-10-23 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Preparation of Selenoinsulin as a Long-Lasting Insulin Analogue.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3A44
 
 | | Crystal structure of HypA in the dimeric form | | 分子名称: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | 著者 | Watanabe, S, Arai, T, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2009-06-30 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
5AYK
 
 | | Crystal structure of ERdj5 form I | | 分子名称: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, DnaJ homolog subfamily C member 10 | | 著者 | Watanabe, S, Maegawa, K, Inaba, K. | | 登録日 | 2015-08-22 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | 分子名称: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | 登録日 | 2012-11-27 | | 公開日 | 2013-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
8IBX
 
 | | Structure of R2 with 3'UTR and DNA in unwinding state | | 分子名称: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | 著者 | Deng, P, Tan, S, Wang, J, Liu, J.J. | | 登録日 | 2023-02-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
 | | Structure of R2 with 3'UTR and DNA in binding state | | 分子名称: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | 著者 | Deng, P, Tan, S, Wang, J, Liu, J.J. | | 登録日 | 2023-02-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
 | | Structure of R2 with 5'ORF and 3'UTR | | 分子名称: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | 著者 | Deng, P, Tan, S, Wang, J, Liu, J.J. | | 登録日 | 2023-02-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
 | | Structure of R2 with 5'ORF | | 分子名称: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | 著者 | Deng, P, Tan, S, Wang, J, Liu, J.J. | | 登録日 | 2023-02-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
1SRS
 
 | |
6ISC
 
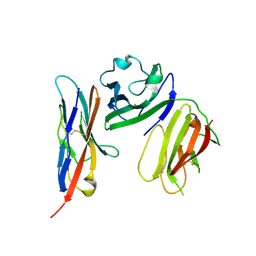 | | complex structure of mCD226-ecto and hCD155-D1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | 著者 | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | 登録日 | 2018-11-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISA
 
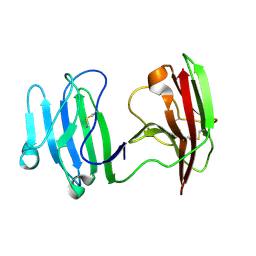 | | mCD226 | | 分子名称: | CD226 antigen | | 著者 | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | 登録日 | 2018-11-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISB
 
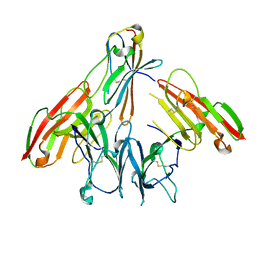 | | crystal structure of human CD226 | | 分子名称: | CD226 antigen | | 著者 | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | 登録日 | 2018-11-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2019-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J14
 
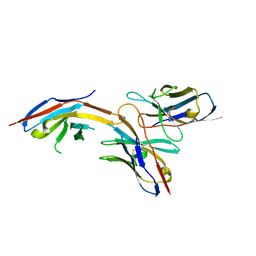 | | Complex structure of GY-14 and PD-1 | | 分子名称: | GY-14 heavy chain V fragment, GY-14 light chain V fragment, Programmed cell death protein 1 | | 著者 | Chen, D, Tan, S, Whang, H, Zhang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2018-12-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
4R8P
 
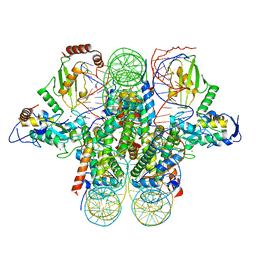 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | 分子名称: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | 著者 | McGinty, R.K, Henrici, R.C, Tan, S. | | 登録日 | 2014-09-02 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.2846 Å) | | 主引用文献 | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | 分子名称: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | 著者 | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | 登録日 | 2016-08-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.206 Å) | | 主引用文献 | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
2XHB
 
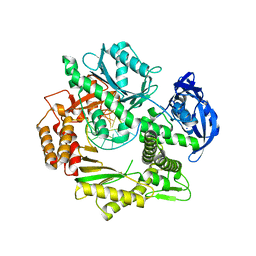 | | Crystal structure of DNA polymerase from Thermococcus gorgonarius in complex with hypoxanthine-containing DNA | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', DNA POLYMERASE, HYPOXANTHINE-CONTAINING DNA, ... | | 著者 | Killelea, T, Ghosh, S, Tan, S.S, Heslop, P, Firbank, S.J, Kool, E.T, Connolly, B.A. | | 登録日 | 2010-06-14 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Probing the Interaction of Archaeal DNA Polymerases with Deaminated Bases Using X-Ray Crystallography and Non-Hydrogen Bonding Isosteric Base Analogues.
Biochemistry, 49, 2010
|
|
6CW2
 
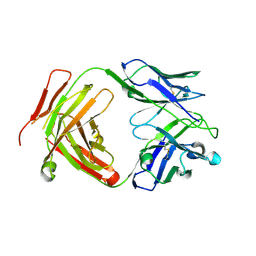 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | 分子名称: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | 著者 | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | 登録日 | 2018-03-29 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CW3
 
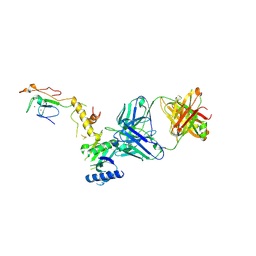 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | 分子名称: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | 著者 | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | 登録日 | 2018-03-29 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-03-04 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7C0D
 
 | |
3MVD
 
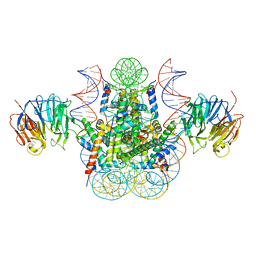 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | 登録日 | 2010-05-04 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
1NH2
 
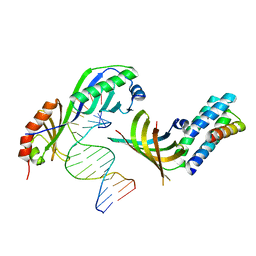 | | Crystal structure of a yeast TFIIA/TBP/DNA complex | | 分子名称: | 5'-D(*GP*TP*TP*TP*TP*AP*TP*AP*TP*AP*CP*AP*TP*AP*CP*A)-3', 5'-D(*TP*GP*TP*AP*(5IU)P*GP*TP*AP*TP*AP*(5IU)P*AP*AP*AP*AP*C)-3', Transcription initiation factor IIA large chain, ... | | 著者 | Bleichenbacher, M, Tan, S, Richmond, T.J. | | 登録日 | 2002-12-18 | | 公開日 | 2003-10-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes.
J.Mol.Biol., 332, 2003
|
|
