5VY2
 
 | |
5W6X
 
 | |
7TDZ
 
 | |
8QC0
 
 | |
8PQQ
 
 | |
8RH3
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to Gemcitabine | | 分子名称: | (2~{R},3~{R})-4,4-bis(fluoranyl)-2-(hydroxymethyl)oxolan-3-ol, Nucleoside 2-deoxyribosyltransferase | | 著者 | Tang, P, Harding, C.J, Czekster, C.M. | | 登録日 | 2023-12-14 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
8PQP
 
 | |
8PQR
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to DAD_Immucillin-H | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Nucleoside 2-deoxyribosyltransferase | | 著者 | Tang, P, Harding, C.J, Czekster, C.M. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.586 Å) | | 主引用文献 | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
8PQT
 
 | |
8PQS
 
 | |
5I8U
 
 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | 分子名称: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | 登録日 | 2016-02-19 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
5WJI
 
 | |
5W6Z
 
 | |
5CY2
 
 | |
4GD8
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA3-53 | | 分子名称: | (2S,3R)-4-(2-aminoethylcarbamoyloxy)-2-[(2-methanoylindolizin-3-yl)amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | 著者 | Pattanaik, P, van den Akker, F. | | 登録日 | 2012-07-31 | | 公開日 | 2013-07-31 | | 最終更新日 | 2013-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
1QHG
 
 | | STRUCTURE OF DNA HELICASE MUTANT WITH ADPNP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HELICASE PCRA, MAGNESIUM ION | | 著者 | Soultanas, P, Dillingham, M.S, Velankar, S.S, Wigley, D.B. | | 登録日 | 1999-05-14 | | 公開日 | 1999-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase.
J.Mol.Biol., 290, 1999
|
|
1QHH
 
 | | STRUCTURE OF DNA HELICASE WITH ADPNP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (PCRA (SUBUNIT)) | | 著者 | Soultanas, P, Dillingham, M.S, Velankar, S.S, Wigley, D.B. | | 登録日 | 1999-05-14 | | 公開日 | 1999-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase.
J.Mol.Biol., 290, 1999
|
|
7DJR
 
 | |
6B09
 
 | |
3D4F
 
 | | SHV-1 beta-lactamase complex with LN1-255 | | 分子名称: | (3R)-4-{[(3,4-dihydroxyphenyl)acetyl]oxy}-N-(2-formylindolizin-3-yl)-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | 著者 | Pattanaik, P, Bethel, C.R, Hujer, A.M, Bonomo, R.A, Buynak, J.D, van den Akker, F. | | 登録日 | 2008-05-14 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Strategic Design of an Effective {beta}-Lactamase Inhibitor: LN-1-255, A 6-ALKYLIDENE-2'-SUBSTITUTED PENICILLIN SULFONE
J.Biol.Chem., 284, 2009
|
|
6CDU
 
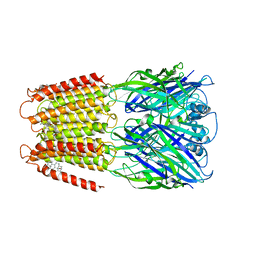 | | Crystal structure of a chimeric human alpha1GABAA receptor in complex with alphaxalone | | 分子名称: | (3a,5a)-3-Hydroxypregnane-11,20-dione, chimeric alpha1GABAA receptor | | 著者 | Chen, Q, Arjunan, P, Cohen, A.E, Xu, Y, Tang, P. | | 登録日 | 2018-02-09 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Structural basis of neurosteroid anesthetic action on GABAAreceptors.
Nat Commun, 9, 2018
|
|
6IZT
 
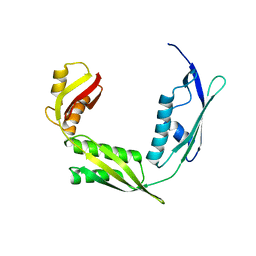 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6J09
 
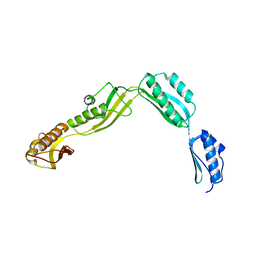 | | Crystal structure of Haemophilus Influenzae BamA POTRA1-4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-21 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
1K4I
 
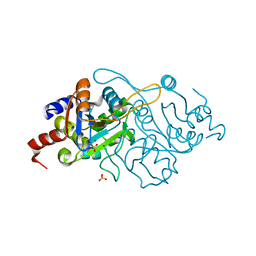 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with two Magnesium ions | | 分子名称: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, MAGNESIUM ION, SULFATE ION | | 著者 | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | 登録日 | 2001-10-08 | | 公開日 | 2002-03-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
