8FWF
 
 | | Crystal structure of Apo form Fab235 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FXJ
 
 | | Crystal structure of Fab460 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab460, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FYM
 
 | | Crystal structure of Fab235 in complex with MPER peptide | | Descriptor: | ALA-SER-LEU-TRP-ASN-TRP-PHE-ASN-ILE-THR-ASN-TRP-LEU-TRP-TYR-ILE-LYS-LYS-LYS, CHLORIDE ION, Fab235, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
3IUV
 
 | | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3 | | Descriptor: | uncharacterized TetR family protein | | Authors: | Tan, K, Cuff, M, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3
To be Published
|
|
3HCZ
 
 | | The crystal structure of a domain of possible thiol-disulfide isomerase from Cytophaga hutchinsonii ATCC 33406. | | Descriptor: | FORMIC ACID, Possible thiol-disulfide isomerase, SULFATE ION | | Authors: | Tan, K, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a domain of possible thiol-disulfide isomerase from Cytophaga hutchinsonii ATCC 33406.
To be Published
|
|
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
3GNJ
 
 | | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB | | Descriptor: | Thioredoxin domain protein | | Authors: | Tan, K, Volkart, L, Gu, M, Kinney, J.N, Babnigg, G, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB
To be Published
|
|
3NZE
 
 | | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1. | | Descriptor: | CALCIUM ION, Putative transcriptional regulator, sugar-binding family | | Authors: | Tan, K, Zhang, R, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1.
To be Published
|
|
3ON3
 
 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
3PN8
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Putative phospho-beta-glucosidase, ... | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutants UA159
To be Published
|
|
8FZ2
 
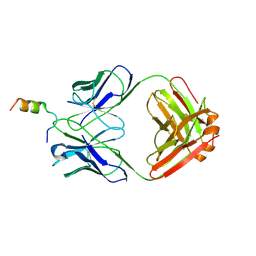 | | Crystal structure of Fab460 in complex with MPER peptide | | Descriptor: | Fab460, H chain, L chain, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3RGL
 
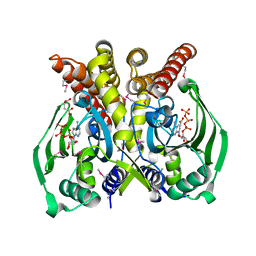 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ADENOSINE-5'-TRIPHOSPHATE, GLYCINE, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine.
To be Published
|
|
3QWT
 
 | |
3OM8
 
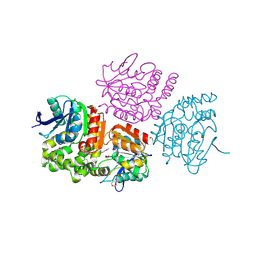 | | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable hydrolase | | Authors: | Tan, K, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01
To be Published
|
|
6X1L
 
 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
3OLQ
 
 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
6XOA
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation
To Be Published
|
|
3Q3C
 
 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3Q13
 
 | | The Structure of the Ca2+-binding, Glycosylated F-spondin Domain of F-spondin, A C2-domain Variant from Extracellular Matrix | | Descriptor: | ACETATE ION, CALCIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Lawler, J. | | Deposit date: | 2010-12-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the Ca2+-binding, glycosylated F-spondin domain of F-spondin - A C2-domain variant in an extracellular matrix protein.
Bmc Struct.Biol., 11, 2011
|
|
3PNN
 
 | |
3PMM
 
 | | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | FORMIC ACID, IMIDAZOLE, Putative cytoplasmic protein | | Authors: | Tan, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3PHA
 
 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
3QZ6
 
 | |
3QZ3
 
 | | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Ferritin | | Authors: | Tan, K, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
