1XN3
 
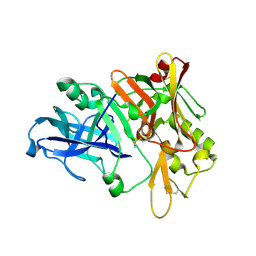 | | Crystal structure of Beta-secretase bound to a long inhibitor with additional upstream residues. | | Descriptor: | Beta-secretase 1, Peptidic inhibitor | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
1XS7
 
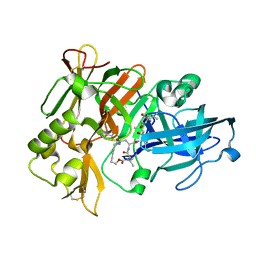 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2I33
 
 | |
2I34
 
 | |
1Y57
 
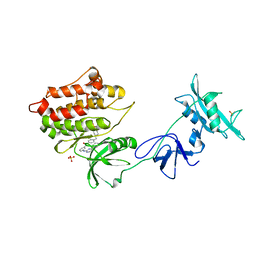 | | Structure of unphosphorylated c-Src in complex with an inhibitor | | Descriptor: | 4-[(4-METHYLPIPERAZIN-1-YL)METHYL]-N-{3-[(4-PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]PHENYL}BENZAMIDE, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P.W, Jahnke, W, Fabbro, D, Liebetanz, J, Meyer, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structure of a c-Src Complex in an Active Conformation Suggests Possible Steps in c-Src Activation
Structure, 13, 2005
|
|
2AY0
 
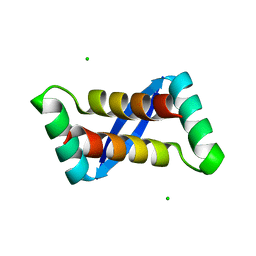 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
1URW
 
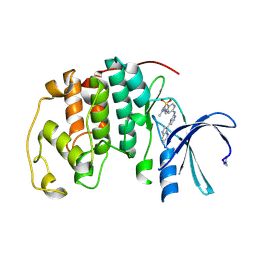 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2OYV
 
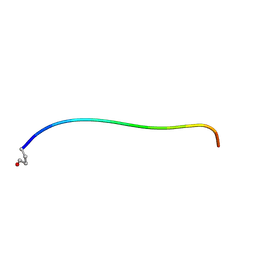 | | Neurotensin in DPC micelles | | Descriptor: | neurotensin | | Authors: | Monti, J.P, Coutant, J, Curmi, P.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Biochemistry, 46, 2007
|
|
2OYW
 
 | | Neurotensin in TFE:H2O (80:20) | | Descriptor: | neurotensin | | Authors: | Monti, J.P, Coutant, J, Curmi, P.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Biochemistry, 46, 2007
|
|
4ZUK
 
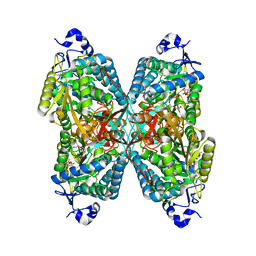 | | Structure ALDH7A1 complexed with NAD+ | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
2NAR
 
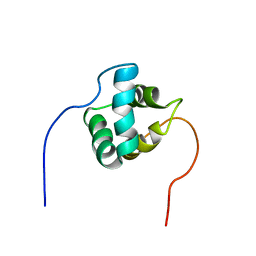 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
1VKG
 
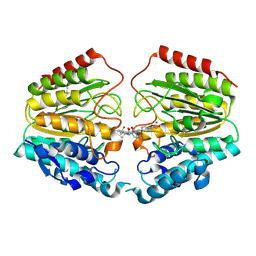 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1J3I
 
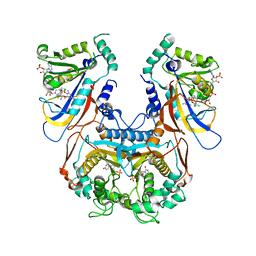 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with WR99210, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
4DPD
 
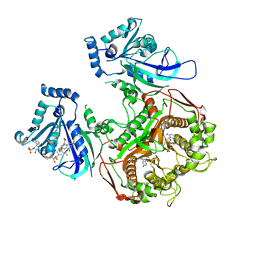 | | WILD TYPE PLASMODIUM FALCIPARUM DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE (PfDHFR-TS), DHF COMPLEX, NADP+, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, DIHYDROFOLIC ACID, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1J3K
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with WR99210, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
1J3J
 
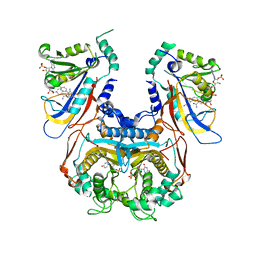 | | Double mutant (C59R+S108N) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with pyrimethamine, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
4BHI
 
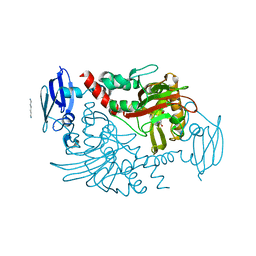 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1,1,1,2-Tetramethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1,1,1,2-TETRAMETHYLHYDRAZIN-1-IUM-2-YL)PROPANOATE, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BGK
 
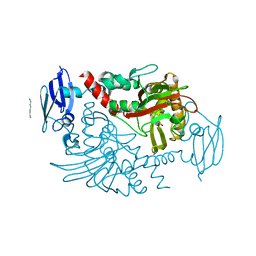 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with (3-(Trimethylammonio)propyl)phosphinate | | Descriptor: | GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, N-OXALYLGLYCINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
2RU8
 
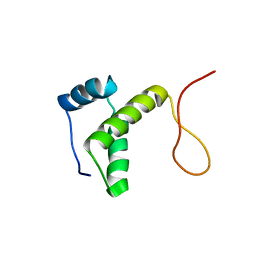 | | DnaT C-terminal domain | | Descriptor: | Primosomal protein 1 | | Authors: | Abe, Y, Tani, J, Fujiyama, S, Urabe, M, Sato, K, Aramaki, T, Katayama, T, Ueda, T. | | Deposit date: | 2014-01-29 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of the primosome protein DnaT-functional structures for homotrimerization, dissociation of ssDNA from the PriB·ssDNA complex, and formation of the DnaT·ssDNA complex.
Febs J., 281, 2014
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | Authors: | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
5Y0V
 
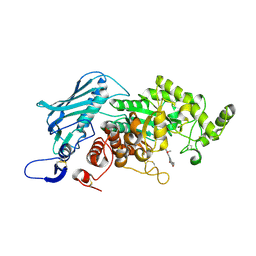 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with berberine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERBERINE, ... | | Authors: | Duan, Y.W, Liu, T, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
4C5W
 
 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 4-(Ethyldimethylammonio)butanoate | | Descriptor: | GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, N-OXALYLGLYCINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BGM
 
 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-Carboxy-N-(2-fluoroethyl)-N,N-dimethylpropan-1- aminium chloride | | Descriptor: | 3-Carboxy-N-(2-fluoroethyl)-N,N-dimethylpropan-1-aminium, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4DSH
 
 | | Crystal structure of reduced UDP-Galactopyranose mutase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Biochemistry, 51, 2012
|
|
4BEZ
 
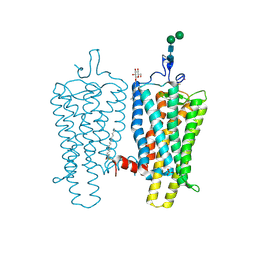 | | Night blindness causing G90D rhodopsin in the active conformation | | Descriptor: | ACETATE ION, PALMITIC ACID, RHODOPSIN, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
