6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | 分子名称: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | 著者 | Costanzi, E, Sormanni, P, Ricagno, S. | | 登録日 | 2020-05-22 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | 分子名称: | DNA polymerase sliding clamp A, ZINC ION | | 著者 | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | 登録日 | 2003-04-28 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
1VE0
 
 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | 分子名称: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | 著者 | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | 登録日 | 2004-03-26 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
9IIE
 
 | | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Destan, E, DeMirci, H. | | 登録日 | 2024-06-20 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
7TV9
 
 | | HUMAN COMPLEMENT COMPONENT C3B IN COMPLEX WITH APL-1030 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APL-1030 Nanofitin, Complement C3 beta chain, ... | | 著者 | Fontano, E, Nadupalli, A, Lakshminarasimhan, D, White, A, Garlish, J, Cinier, M, Chevrel, A, Perrocheau, A, Eyerman, D, Orme, M, Kitten, O, Scheibler, L. | | 登録日 | 2022-02-04 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Discovery of APL-1030, a Novel, High-Affinity Nanofitin Inhibitor of C3-Mediated Complement Activation.
Biomolecules, 12, 2022
|
|
1EIE
 
 | |
1EID
 
 | |
1EIC
 
 | |
1FS3
 
 | |
7B3E
 
 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-11-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7ALI
 
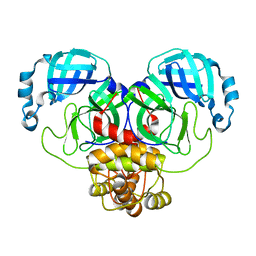 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
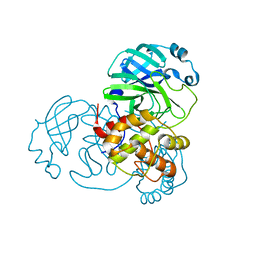 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8A2C
 
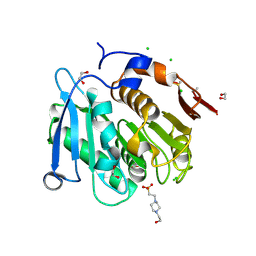 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
8OKM
 
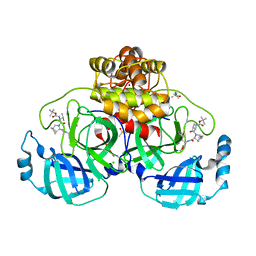 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKL
 
 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKN
 
 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
7BFB
 
 | | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | 1,2-ETHANEDIOL, Main Protease, N-phenyl-2-selanylbenzamide, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-01-02 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
7BGP
 
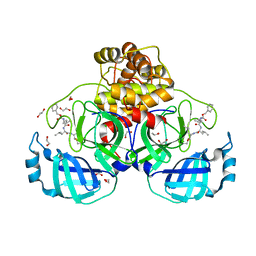 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-01-08 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BE7
 
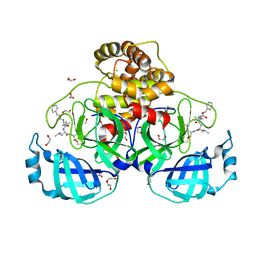 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-12-22 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BB2
 
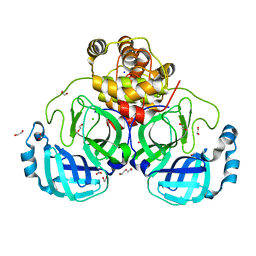 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-12-16 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8A5W
 
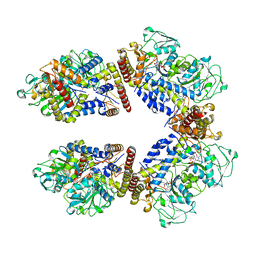 | | Crystal structure of the human phosphoserine aminotransferase (PSAT) in complex with O-phosphoserine | | 分子名称: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, PHOSPHOSERINE, ... | | 著者 | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | 登録日 | 2022-06-16 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
8A5V
 
 | | Crystal structure of the human phosposerine aminotransferase (PSAT) | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase, SULFATE ION | | 著者 | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | 登録日 | 2022-06-16 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
8B4U
 
 | | The crystal structure of PET46, a PETase enzyme from Candidatus bathyarchaeota | | 分子名称: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Applegate, V, Schumacher, J, Smits, S.H.J. | | 登録日 | 2022-09-21 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate.
Commun Chem, 6, 2023
|
|
8P86
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-31 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
