2SOD
 
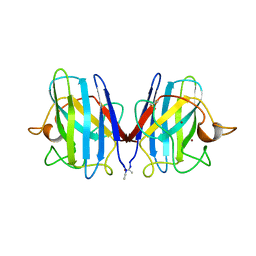 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
1AKZ
 
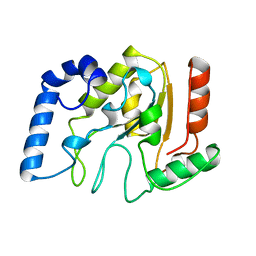 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
8VGY
 
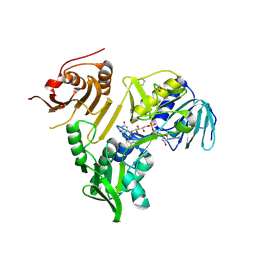 | | Crystal structure of human apoptosis-inducing factor (AIF) bound to the fused N-terminal domain of CHCHD4 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis-inducing factor 1, mitochondrial,Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Brosey, C.A, Tainer, J.A. | | Deposit date: | 2023-12-29 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
Embo J., 44, 2025
|
|
6O9X
 
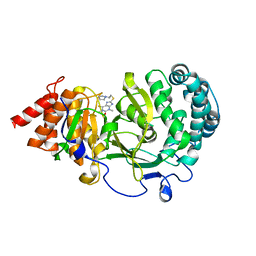 | | Structure of human PARG complexed with JA2-4 | | Descriptor: | 1,3-dimethyl-8-{[2-(pyrrolidin-1-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,9-tetrahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OA1
 
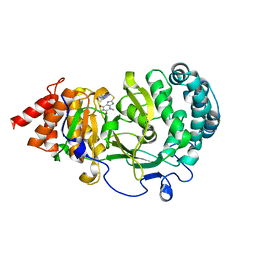 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
1EMH
 
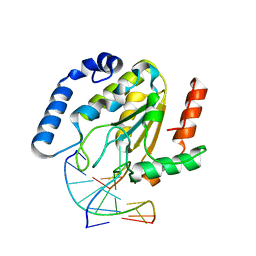 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO UNCLEAVED SUBSTRATE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(P2U)P*AP*TP*CP*TP*T)-3'), URACIL-DNA GLYCOSYLASE | | Authors: | Parikh, S.S, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EM1
 
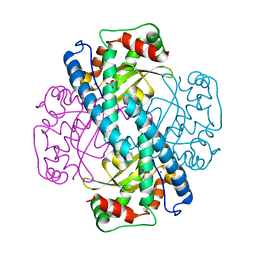 | | X-RAY CRYSTAL STRUCTURE FOR HUMAN MANGANESE SUPEROXIDE DISMUTASE, Q143A | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE, SULFATE ION | | Authors: | Leveque, V, Stroupe, M.E, Lepock, J.R, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2000-03-14 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Multiple replacements of glutamine 143 in human manganese superoxide dismutase: effects on structure, stability, and catalysis.
Biochemistry, 39, 2000
|
|
6VBH
 
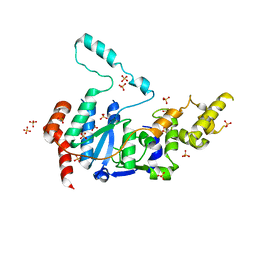 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3L40
 
 | |
3GVA
 
 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
8GJ9
 
 | | RAD51C N-terminal domain | | Descriptor: | RAD51C, ZINC ION | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJA
 
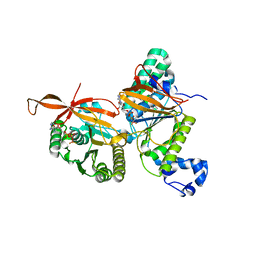 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJ8
 
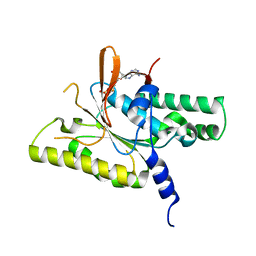 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
3UEK
 
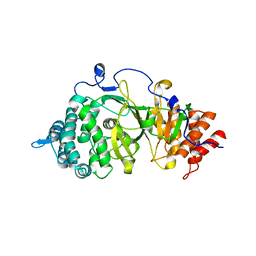 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | Descriptor: | Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEL
 
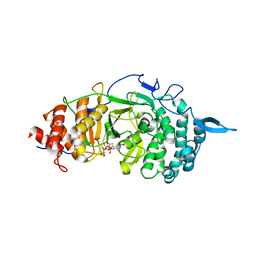 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UMV
 
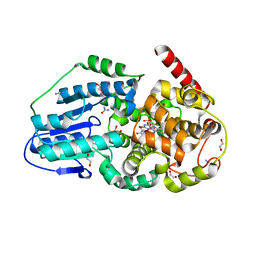 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
1M8H
 
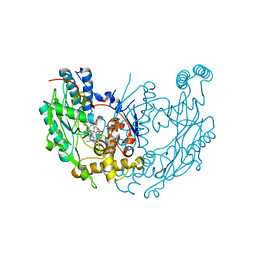 | | inducible nitric oxide synthase with 6-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M9K
 
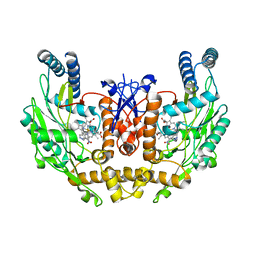 | | Human Endothelial Nitric Oxide Synthase with 7-Nitroindazole Bound | | Descriptor: | 7-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M9A
 
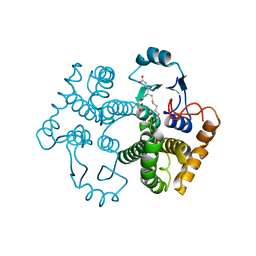 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9Q
 
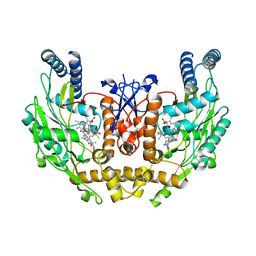 | | human endothelial nitric oxide synthase with 5-nitroindazole bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-NITROINDAZOLE, ISOPROPYL ALCOHOL, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M8D
 
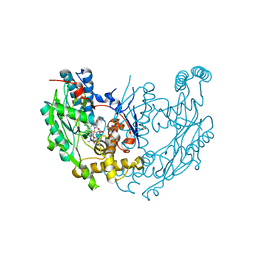 | | inducible nitric oxide synthase with Chlorzoxazone bound | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORZOXAZONE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M9T
 
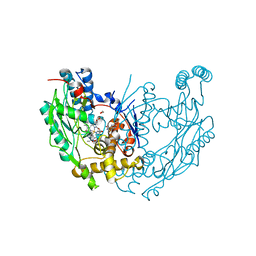 | | Inducible Nitric Oxide Synthase with 3-Bromo-7-Nitroindazole bound | | Descriptor: | 1,2-ETHANEDIOL, 3-BROMO-7-NITROINDAZOLE, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-09-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
8D3G
 
 | |
8D3J
 
 | |
