3L2P
 
 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
6ES3
 
 | | Structure of CDX2-DNA(TCG) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6ES2
 
 | | Structure of CDX2-DNA(CAA) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6M7L
 
 | | Complex of OxyA with the X-domain from GPA biosynthesis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | Authors: | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.648297 Å) | | Cite: | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
2ABK
 
 | |
1QNM
 
 | | HUMAN MANGANESE SUPEROXIDE DISMUTASE MUTANT Q143N | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the active site of human manganese superoxide dismutase: the role of glutamine 143.
Biochemistry, 37, 1998
|
|
1QUM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA | | Descriptor: | 5'-D(*(3DR)P*CP*GP*AP*CP*GP*A)-3', 5'-D(*CP*GP*TP*CP*C)-3', 5'-D(*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*G)-3', ... | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1QTW
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
2AWJ
 
 | | GFP R96M pre-cyclized intermediate in chromophore formation | | Descriptor: | MAGNESIUM ION, green-fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
1QYF
 
 | | Crystal structure of matured green fluorescent protein R96A variant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-10 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1QYQ
 
 | | Crystal Structure of the cyclized S65G Y66G GFP variant | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
8D3J
 
 | |
8D3E
 
 | |
8D3O
 
 | |
8D3I
 
 | |
8D3N
 
 | |
8D3K
 
 | |
8D3G
 
 | |
8D3H
 
 | |
6VBH
 
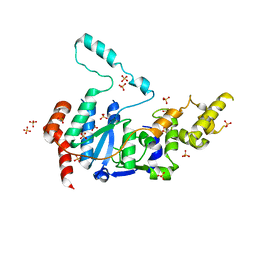 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VBA
 
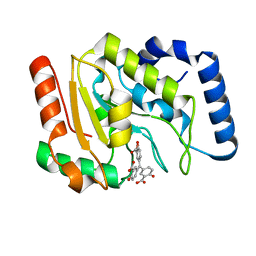 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6X1Z
 
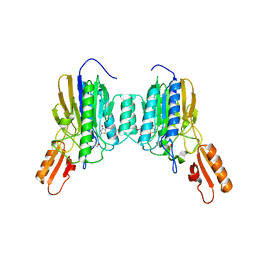 | | Mre11 dimer in complex with small molecule modulator PFMJ | | Descriptor: | (5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6WH2
 
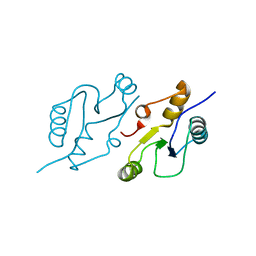 | | Structure of the C-terminal BRCT domain of human XRCC1 | | Descriptor: | X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
6WH1
 
 | | Structure of the complex of human DNA ligase III-alpha and XRCC1 BRCT domains | | Descriptor: | DNA ligase 3 alpha, X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
7Q3O
 
 | |
