3QQD
 
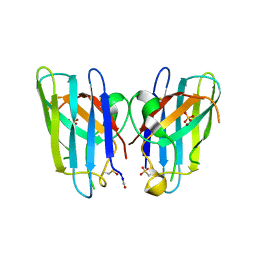 | | Human SOD1 H80R variant, P212121 crystal form | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Schirf, V, Demeler, B, Carroll, M.C, Culotta, V.C, Hart, P.J. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R.
Biochemistry, 49, 2010
|
|
6CBY
 
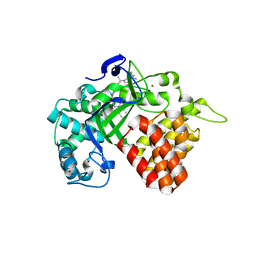 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF9975 | | Descriptor: | N-lysine methyltransferase SMYD2, ZINC ION, [3-(4-amino-6-methyl-1H-imidazo[4,5-c]pyridin-1-yl)-3-methylazetidin-1-yl][1-({1-[(1R)-cyclohept-2-en-1-yl]piperidin-4-yl}methyl)-1H-pyrrol-3-yl]methanone | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective, Small-Molecule Co-Factor Binding Site Inhibition of a Su(var)3-9, Enhancer of Zeste, Trithorax Domain Containing Lysine Methyltransferase.
J.Med.Chem., 62, 2019
|
|
3KBE
 
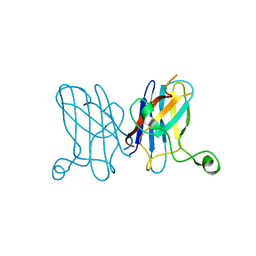 | | Metal-free C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
7TAB
 
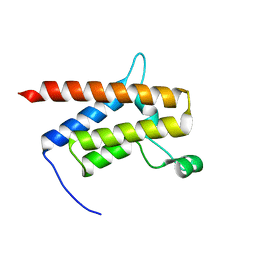 | | G-925 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 2-(6-amino-5-phenylpyridazin-3-yl)phenol, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
7TD9
 
 | | G-059 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 4-phenyl-5H-pyridazino[4,3-b]indol-3-amine, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
1LAP
 
 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
5CU9
 
 | | CANDIDA ALBICANS SUPEROXIDE DISMUTASE 5 (SOD5), APO | | Descriptor: | Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Waninger-Saroni, J.J, Taylor, A.B, Hart, P.J, Galaleldeen, A. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | CANDIDA ALBICANS SUPEROXIDE DISMUTASE 5 (SOD5), APO
To Be Published
|
|
5K71
 
 | | apo Dbr1 | | Descriptor: | RNA lariat debranching enzyme, putative, SULFATE ION | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
5K73
 
 | | as-isolated Dbr1 with Fe(II) and Zn(II) | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
3Q6E
 
 | | Human insulin in complex with cucurbit[7]uril | | Descriptor: | Insulin A chain, Insulin B chain, cucurbit[7]uril | | Authors: | Chinai, J.M, Taylor, A.B, Hargreaves, N.D, Ryno, L.M, Morris, C.A, Hart, P.J, Urbach, A.R. | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular recognition of insulin by a synthetic receptor.
J.Am.Chem.Soc., 133, 2011
|
|
5KBL
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110Q Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5K78
 
 | | Dbr1 in complex with 16-mer branched RNA | | Descriptor: | FE (II) ION, RNA lariat debranching enzyme, putative, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
5KBM
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), D113N Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5KBK
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110A Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
2VYN
 
 | | Structure of E.Coli GAPDH Rat Sperm GAPDH heterotetramer | | Descriptor: | FORMIC ACID, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Frayne, J, Taylor, A, Hall, L, Hadfield, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Insoluble Rat Sperm Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) Via Heterotetramer Formation with Escherichia Coli Gapdh Reveals Target for Contraceptive Design.
J.Biol.Chem., 284, 2009
|
|
2VYV
 
 | | Structure of E.Coli GAPDH Rat Sperm GAPDH heterotetramer | | Descriptor: | FORMIC ACID, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Frayne, J, Taylor, A, Hall, L, Hadfield, A. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of Insoluble Rat Sperm Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) Via Heterotetramer Formation with Escherichia Coli Gapdh Reveals Target for Contraceptive Design.
J.Biol.Chem., 284, 2009
|
|
3KBF
 
 | | C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (II) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
6CBX
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-lysine methyltransferase SMYD2, ... | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497
to be published
|
|
5K77
 
 | | Dbr1 in complex with 7-mer branched RNA | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8CZM
 
 | |
8CZN
 
 | |
8DZK
 
 | | Dbr1 in complex with 5-mer cleavage product | | Descriptor: | FE (II) ION, RNA (5'-R(P*(G46)P*UP*GP*UP*U)-3'), RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B. | | Deposit date: | 2022-08-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product.
Biochemistry, 61, 2022
|
|
2N1L
 
 | | Solution structure of the BCOR PUFD | | Descriptor: | BCL-6 corepressor | | Authors: | Wong, S.J, Gearhart, M.D, Ha, D.J, Corcoran, C.M, Diaz, V, Taylor, A.B, Schirf, V, Ilangovan, U, Hinck, A.P, Demeler, B, Hart, J, Bardwell, V.J, Kim, C.A. | | Deposit date: | 2015-04-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the hierarchical assembly of the core of PRC1.1
To be Published
|
|
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
