3V0S
 
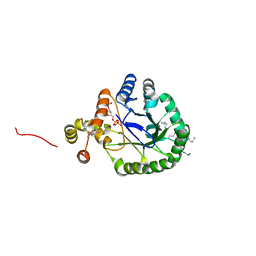 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
8EOP
 
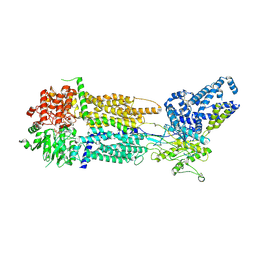 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
3V71
 
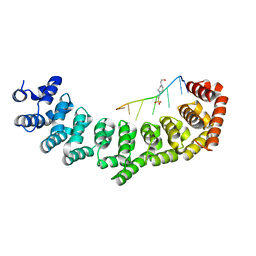 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
8SPO
 
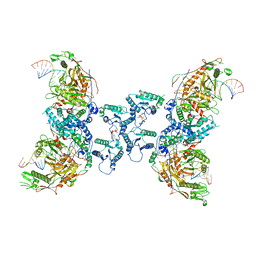 | | Tetramerized activation of MapSPARTA bound with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain-containing protein, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8SQU
 
 | | Monomeric MapSPARTA bound with guide RNA and target DNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
3UNY
 
 | | Bacillus cereus phosphopentomutase T85E variant soaked with glucose 1,6-bisphosphate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UYI
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UZX
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and epiandrosterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 3-oxo-5-beta-steroid 4-dehydrogenase, 5-ALPHA-ANDROSTANE-3-BETA,17BETA-DIOL, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
3UZZ
 
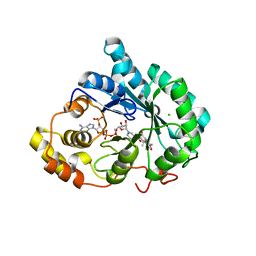 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and delta4-androstenedione | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
4BRV
 
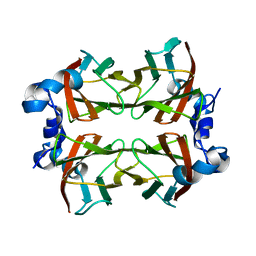 | | Superoxide reductase (Neelaredoxin) from Ignicoccus hospitalis E23A | | Descriptor: | DESULFOFERRODOXIN, FERROUS IRON-BINDING REGION, FE (III) ION | | Authors: | Romao, C.V, Matias, P.M, Pinho, F.G, Sousa, C.M, Barradas, A.R, Pinto, A.F, Teixeira, M, Bandeiras, T.M. | | Deposit date: | 2013-06-05 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a Natural Sor Mutant
To be Published
|
|
3V0T
 
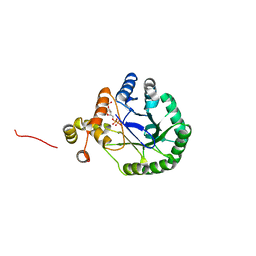 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
4BMK
 
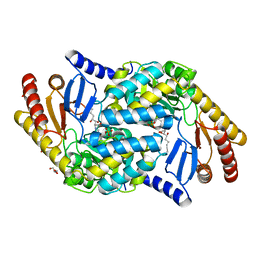 | | Serine Palmitoyltransferase K265A from S. paucimobilis with bound PLP- Myriocin Aldimine | | Descriptor: | Decarboxylated Myriocin, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wadsworth, J.M, Clarke, D.J, McMahon, S.A, Beattie, A.E, Lowther, J, Dunn, T.M, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin.
J.Am.Chem.Soc., 135, 2013
|
|
4BRJ
 
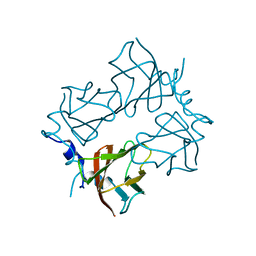 | | Superoxide reductase (Neelaredoxin) from Ignicoccus hospitalis T24K | | Descriptor: | DESULFOFERRODOXIN, FERROUS IRON-BINDING REGION, FE (III) ION | | Authors: | Romao, C.V, Matias, P.M, Pinho, F.G, Sousa, C.M, Barradas, A.R, Pinto, A.F, Teixeira, M, Bandeiras, T.M. | | Deposit date: | 2013-06-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Natural Sor Mutant
To be Published
|
|
4CZC
 
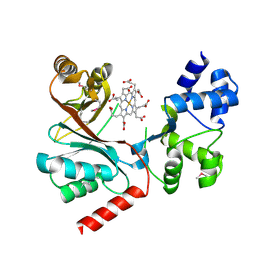 | | Crystal structure of the siroheme decarboxylase NirDL in co-complex with iron-uroporphyrin III analogue | | Descriptor: | Fe(III) Uroporphyrin, NIRD-LIKE PROTEIN | | Authors: | Schmelz, S, Kriegler, T.M, Haufschildt, K, Layer, G, Heinz, D.W. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Siroheme Decarboxylase in Complex with Iron-Uroporphyrin III Reveals Two Essential Histidine Residues
J.Mol.Biol., 426, 2014
|
|
3V74
 
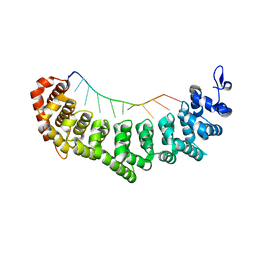 | | crystal structure of FBF-2 in complex with gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*CP*AP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Wang, Y, Qiu, C, Kershner, A, Holley, C.P, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
8BEJ
 
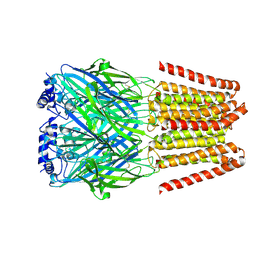 | | GABA-A receptor a5 homomer - a5V3 - APO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHM
 
 | | GABA-A receptor a5 homomer - a5V3 - DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, methyl 4-ethyl-6,7-dimethoxy-9H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHB
 
 | | GABA-A receptor a5 homomer - a5V3 - RO154513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl 8-[(azanylidene-$l^{4}-azanylidene)amino]-5-methyl-6-oxidanylidene-4~{H}-imidazo[1,5-a][1,4]benzodiazepine-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y, Wahid, A.A. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHI
 
 | | GABA-A receptor a5 homomer - a5V3 - RO5211223 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, [1,1-bis(oxidanylidene)-1,4-thiazinan-4-yl]-[6-[[5-methyl-3-(6-methylpyridin-3-yl)-1,2-oxazol-4-yl]methoxy]pyridin-3-yl]methanone | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EIN
 
 | |
8BHK
 
 | | GABA-A receptor a5 homomer - a5V3 - Diazepam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Kasaragod, V.B, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EIP
 
 | |
8BHG
 
 | | GABA-A receptor a5 heteromer - a5V2 - Bretazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bretazenil, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHO
 
 | | GABA-A receptor a5 homomer - a5V3 - L655708 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl (7~{S})-15-methoxy-12-oxidanylidene-2,4,11-triazatetracyclo[11.4.0.0^{2,6}.0^{7,11}]heptadeca-1(17),3,5,13,15-pentaene-5-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHQ
 
 | | GABA-A receptor a5 homomer - a5V3 - RO7172670 | | Descriptor: | 2-[[5-methyl-3-(6-methylpyridazin-3-yl)-1,2-oxazol-4-yl]methyl]-5-(5-oxa-2-azaspiro[3.5]nonan-2-yl)pyridazin-3-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
