5ES8
 
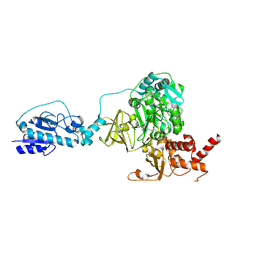 | | Crystal structure of the initiation module of LgrA in the thiolation state | | Descriptor: | Linear gramicidin synthetase subunit A, [(3~{R})-4-[[3-[2-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Reimer, J.M, Aloise, M.N, Schmeing, T.M. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase.
Nature, 529, 2016
|
|
5ES6
 
 | |
5ES5
 
 | |
5EO0
 
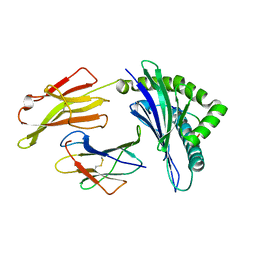 | | Crystal Structure of HLA-B0702-RFL9 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Josephs, T.M, Gras, S, Rossjohn, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-24 | | Last modified: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1-Specific CD8 T Cells Exhibit Limited Cross-Reactivity during Acute Infection.
J Immunol., 196, 2016
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
5ES9
 
 | |
1ITG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
1JQZ
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag. | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-09 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
3A2S
 
 | |
1JT3
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Histidine Tag AND LEU 73 REPLACED BY VAL (L73V) | | Descriptor: | SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
5BMP
 
 | |
5BMN
 
 | | Crystal Structure of APO form of Phosphoglucomutase from Xanthomonas citri | | Descriptor: | MAGNESIUM ION, Phosphoglucomutase | | Authors: | Goto, L.S, Pereira, H.M, Novo Mansur, M.T.M, Brandao-Neto, J. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri.
Biochim.Biophys.Acta, 1864, 2016
|
|
5B6C
 
 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
1K74
 
 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K5F
 
 | | SOLUTION STRUCTURE OF THE S-STYRENE ADDUCT IN THE RAS61 SEQUENCE | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(ABS)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Hennard, C, Finneman, J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-10-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The nonmutagenic (R)- and (S)-beta-(N(6)-adenyl)styrene oxide adducts are oriented in the major groove and show little perturbation to DNA
structure.
Biochemistry, 40, 2001
|
|
1I3K
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
1I3N
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
5BYM
 
 | |
1JT5
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 73 REPLACED BY VAL AND VAL 109 REPLACED BY LEU (L73V/V109L) | | Descriptor: | SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1HQX
 
 | | R308K ARGINASE VARIANT | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Lavulo, L.T, Sossong Jr, T.M, Brigham-Burke, M.R, Doyle, M.L, Cox, J.D, Christianson, D.W, Ash, D.E. | | Deposit date: | 2000-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid.
J.Biol.Chem., 276, 2001
|
|
1HN4
 
 | | PROPHOSPHOLIPASE A2 DIMER COMPLEXED WITH MJ33, SULFATE, AND CALCIUM | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PROPHOSPHOLIPASE A2, ... | | Authors: | Epstein, T.M, Pan, Y.H, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-12-06 | | Release date: | 2001-12-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The basis for k(cat) impairment in prophospholipase A(2) from the anion-assisted dimer structure.
Biochemistry, 40, 2001
|
|
2YP6
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
7NE8
 
 | | Tick salivary protein BSAP1 | | Descriptor: | tick salivary protein BSAP1 | | Authors: | Denisov, S.S, Ippel, J.H, Castoldi, E, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of anticoagulant and anticomplement activity of the tick salivary protein Salp14 and its homologs.
J.Biol.Chem., 297, 2021
|
|
2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
