7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
4EU6
 
 | | Succinyl-CoA:acetate CoA-transferase (AarCH6) in complex with CoA, acetate, and covalent acetylglutamyl anhydride and glutamyl-CoA thioester adducts | | Descriptor: | ACETATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Mullins, E.A, Kappock, T.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal Structures of Acetobacter aceti Succinyl-Coenzyme A (CoA):Acetate CoA-Transferase Reveal Specificity Determinants and Illustrate the Mechanism Used by Class I CoA-Transferases.
Biochemistry, 51, 2012
|
|
1OF3
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module, TmCBM27 | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1PGS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF PNGASE F, A GLYCOSYLASPARAGINASE FROM FLAVOBACTERIUM MENINGOSEPTICUM | | Descriptor: | PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F | | Authors: | Norris, G.E, Stillman, T.J, Anderson, B.F, Baker, E.N. | | Deposit date: | 1994-10-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Structure, 2, 1994
|
|
4JDH
 
 | |
4KW8
 
 | |
7NW2
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-47 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Douangamath, A, Aimon, A, Brandao-Neto, J, Dias, A, Dunnett, L, Gehrtz, P, Gorrie-Stone, T.J, Lukacik, P, Powell, A.J, Skyner, R, Strain-Damerell, C.M, Zaidman, D, London, N, Walsh, M.A, von Delft, F, Covid Moonshot Consortium | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 M pro inhibitor.
Cell Chem Biol, 28, 2021
|
|
1OH4
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
4JNB
 
 | | Crystal structure of the Catalytic Domain of Human DUSP12 | | Descriptor: | Dual specificity protein phosphatase 12, SULFATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1M41
 
 | | Crystal structure of Escherichia coli alkanesulfonate monooxygenase SsuD at 2.3 A resolution | | Descriptor: | FMNH2-dependent alkanesulfonate monooxygenase | | Authors: | Eichhorn, E, Davey, C.A, Sargent, D.F, Leisinger, T, Richmond, T.J. | | Deposit date: | 2002-07-02 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Alkanesulfonate Monooxygenase SsuD
J.mol.biol., 324, 2002
|
|
7VPW
 
 | |
1FKI
 
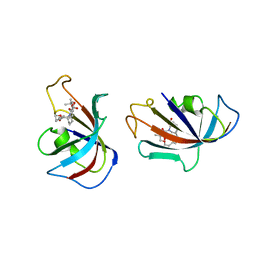 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | (21S)-1AZA-4,4-DIMETHYL-6,19-DIOXA-2,3,7,20-TETRAOXOBICYCLO[19.4.0] PENTACOSANE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
4KV2
 
 | |
4KW9
 
 | |
4KW4
 
 | |
1XW4
 
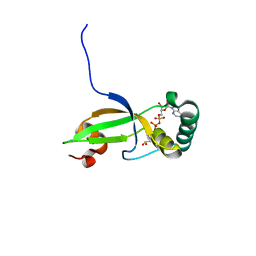 | | Crystal Structure of Human Sulfiredoxin (Srx) in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Sulfiredoxin | | Authors: | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
1FKR
 
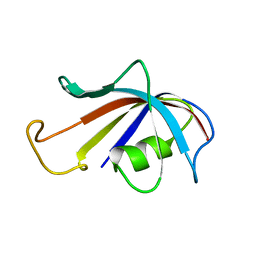 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKG
 
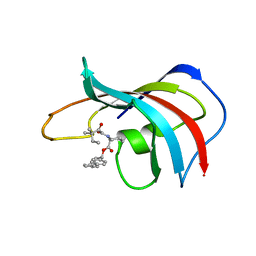 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1,3-DIPHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
4JDK
 
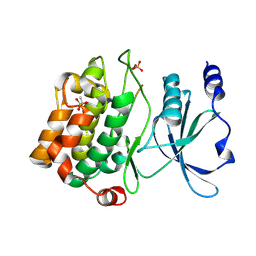 | |
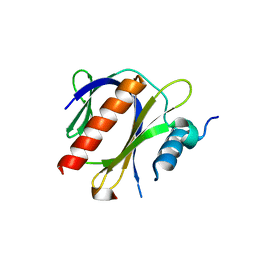
4JIF
 
 | |
1FZL
 
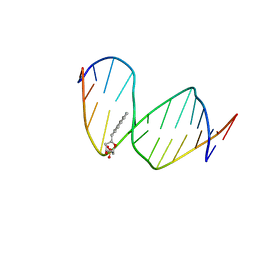 | | DNA WITH PYRENE PAIRED AT ABASIC SITES | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*AP*CP*AP*(PYP)P*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*(3DR)P*TP*GP*TP*TP*TP*GP*TP*G)-3' | | Authors: | Smirnov, S, Matray, T.J, Kool, E.T, de los Santos, C. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Integrity of duplex structures without hydrogen bonding: DNA with pyrene paired at abasic sites
Nucleic Acids Res., 30, 2002
|
|
1EGW
 
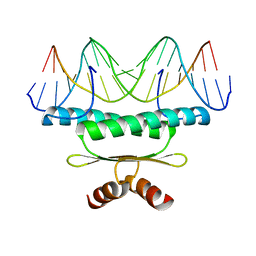 | | CRYSTAL STRUCTURE OF MEF2A CORE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, ... | | Authors: | Santelli, E, Richmond, T.J. | | Deposit date: | 2000-02-17 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of MEF2A core bound to DNA at 1.5 A resolution.
J.Mol.Biol., 297, 2000
|
|
1EJ7
 
 | |
