8JF1
 
 | |
6EFN
 
 | | Structure of a RiPP maturase, SkfB | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Grell, T.A.J, Drennan, C.L. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Structure of a RiPP maturase, SkfB
J.Biol.Chem., 2018
|
|
8JEZ
 
 | |
6EHG
 
 | | complement component C3b in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2CBR
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN I IN COMPLEX WITH A RETINOBENZOIC ACID (AM80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, PROTEIN (CRABP-I) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-12 7310) | | Descriptor: | (2E,4E,6E,8E)-9-(4-hydroxy-2,3,6-trimethylphenyl)-3,7-dimethylnona-2,4,6,8-tetraenoic acid, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3CTI
 
 | |
5BRF
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor HPOP-GlcN | | Descriptor: | 2-deoxy-2-{[3-(4-hydroxyphenyl)propanoyl]amino}-alpha-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRE
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRD
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor BENZ-GlcN | | Descriptor: | 2-(benzoylamino)-2-deoxy-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRH
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor DBT-GlcN | | Descriptor: | 2-deoxy-2-({[(1,1-dioxido-1-benzothiophen-2-yl)methoxy]carbonyl}amino)-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
2REB
 
 | |
1CBR
 
 | |
4MDP
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens in complex with glucose | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6YUR
 
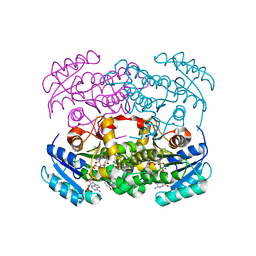 | | Crystal structure of S. aureus FabI inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weinrich, J.D, Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
6YUU
 
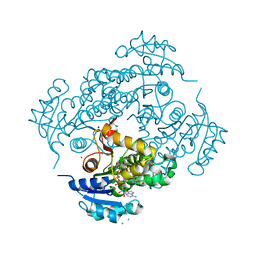 | | Crystal structure of M. tuberculosis InhA inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Weinrich, J.D, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
3CBH
 
 | |
4M3P
 
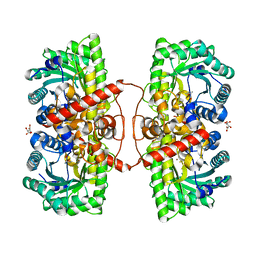 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
3BN3
 
 | | crystal structure of ICAM-5 in complex with aL I domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhang, H, Springer, T.A, Wang, J.-h. | | Deposit date: | 2007-12-13 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | An unusual allosteric mobility of the C-terminal helix of a high-affinity alpha L integrin I domain variant bound to ICAM-5
Mol.Cell, 31, 2008
|
|
1EGN
 
 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2000-02-16 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
6TUQ
 
 | |
4MLS
 
 | | Crystal structure of the SpyTag and SpyCatcher-deltaN1 complex | | Descriptor: | Fibronectin binding protein, SpyTag | | Authors: | Li, L, Fierer, J.O, Rapoport, T.A, Howarth, M. | | Deposit date: | 2013-09-06 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag.
J.Mol.Biol., 426, 2014
|
|
3CAI
 
 | | Crystal structure of Mycobacterium tuberculosis Rv3778c protein | | Descriptor: | POSSIBLE AMINOTRANSFERASE | | Authors: | Covarrubias, A.S, Larsson, A.M, Jones, T.A, Bergfors, T, Mowbray, S.L, Unge, T. | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical function studies of Mycobacterium tuberculosis essential protein Rv3778c
To be published
|
|
6ZEC
 
 | | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Allen, J.D, Watanabe, Y, Crispin, M, Bowden, T.A. | | Deposit date: | 2020-06-16 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insertion of atypical glycans into the tumor antigen-binding site identifies DLBCLs with distinct origin and behavior.
Blood, 138, 2021
|
|
