2EPZ
 
 | | Solution structure of the 4th C2H2 type zinc finger domain of Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Futami, K, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 4th C2H2 type zinc finger domain of Zinc finger protein 28 homolog
To be Published
|
|
2EPV
 
 | | Solution structure of the 20th C2H2 type zinc finger domain of Zinc finger protein 268 | | Descriptor: | ZINC ION, Zinc finger protein 268 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 20th C2H2 type zinc finger domain of Zinc finger protein 268
To be Published
|
|
2EQ1
 
 | | Solution structure of the 9th C2H2 type zinc finger domain of Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 9th C2H2 type zinc finger domain of Zinc finger protein 347
To be Published
|
|
2EPU
 
 | | Solution structure of the secound C2H2 type zinc finger domain of Zinc finger protein 32 | | Descriptor: | ZINC ION, Zinc finger protein 32 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the secound C2H2 type zinc finger domain of Zinc finger protein 32
To be Published
|
|
2EQ2
 
 | | Solution structure of the 16th C2H2 type zinc finger domain of Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 16th C2H2 type zinc finger domain of Zinc finger protein 347
To be Published
|
|
2EQ0
 
 | | Solution structure of the 8th C2H2 type zinc finger domain of Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Masuda, K, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 8th C2H2 type zinc finger domain of Zinc finger protein 347
To be Published
|
|
2EPX
 
 | | Solution structure of the third C2H2 type zinc finger domain of Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third C2H2 type zinc finger domain of Zinc finger protein 28 homolog
To be Published
|
|
2EPT
 
 | | Solution structure of the first C2H2 type zinc finger domain of Zinc finger protein 32 | | Descriptor: | ZINC ION, Zinc finger protein 32 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first C2H2 type zinc finger domain of Zinc finger protein 32
To be Published
|
|
2EQ3
 
 | | Solution structure of the 17th C2H2 type zinc finger domain of Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Futami, K, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 17th C2H2 type zinc finger domain of Zinc finger protein 347
To be Published
|
|
2EPR
 
 | | Solution structure of the secound zinc finger domain of Zinc finger protein 278 | | Descriptor: | POZ-, AT hook-, and zinc finger-containing protein 1, ... | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the secound zinc finger domain of Zinc finger protein 278
To be Published
|
|
3EF4
 
 | | Crystal structure of native pseudoazurin from Hyphomicrobium denitrificans | | Descriptor: | Blue copper protein, COPPER (II) ION, PHOSPHATE ION | | Authors: | Hira, D, Nojiri, M, Suzuki, S. | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Atomic resolution structure of pseudoazurin from the methylotrophic denitrifying bacterium Hyphomicrobium denitrificans: structural insights into its spectroscopic properties
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5H5L
 
 | | Structure of prostaglandin synthase D of Nilaparvata lugens | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, S, Nakagawa, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular structure of a prostaglandin D synthase requiring glutathione from the brown planthopper, Nilaparvata lugens
Biochem. Biophys. Res. Commun., 492, 2017
|
|
2D0W
 
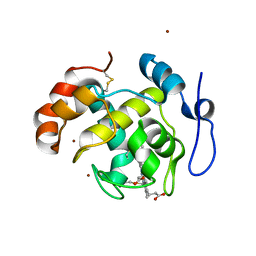 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
2D0V
 
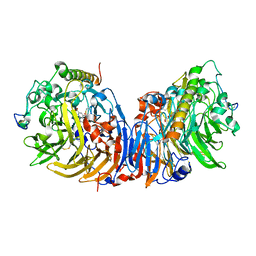 | | Crystal structure of methanol dehydrogenase from Hyphomicrobium denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase large subunit, ... | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-09 | | Release date: | 2006-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
6JT3
 
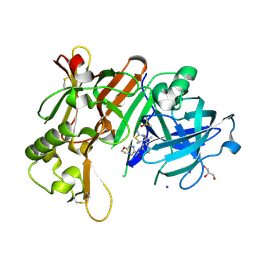 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
3WNM
 
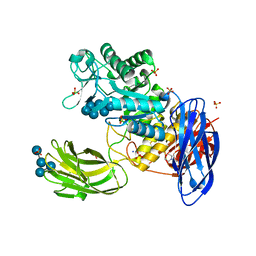 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNL
 
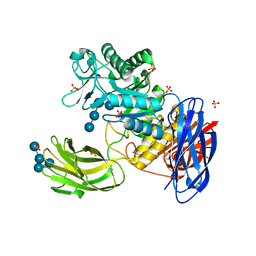 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNN
 
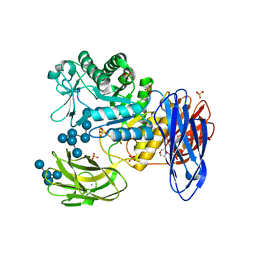 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
6JT4
 
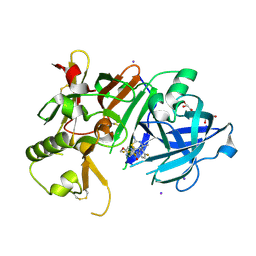 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
7XZS
 
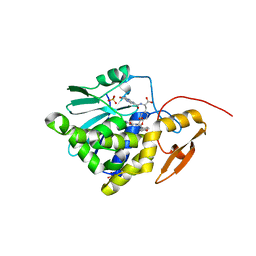 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-tyrosine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZT
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-tyrosine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZW
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-phenylalanine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZU
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-phenylalanine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y02
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-3-(4-fluorophenyl)propanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-fluorophenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
