4H32
 
 | | The crystal structure of the hemagglutinin H17 derived the bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, X, Shi, Y, Lu, X, He, J, Gao, F, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2012-09-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism.
Cell Rep, 3, 2013
|
|
5GJ9
 
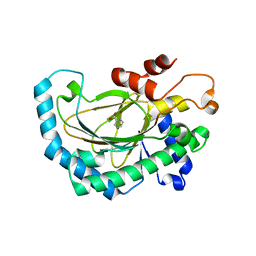 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
5GJA
 
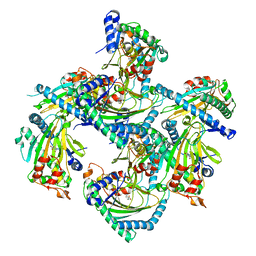 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
5GJ6
 
 | |
5JDB
 
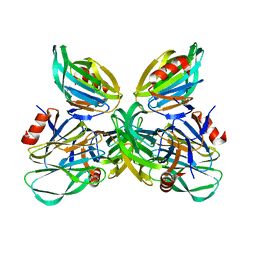 | | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens | | Descriptor: | Outer capsid protein VP4 | | Authors: | Sun, X, Guo, N, Li, D, Zhou, Y, Jin, M, Xie, G, Pang, L, Zhang, Q, Cao, Y, Duan, Z. | | Deposit date: | 2016-04-16 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens.
Virology, 495, 2016
|
|
1FGS
 
 | | FOLYLPOLYGLUTAMATE SYNTHETASE FROM LACTOBACILLUS CASEI | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHETASE, MAGNESIUM ION, PYROPHOSPHATE 2- | | Authors: | Sun, X, Bognar, A, Baker, E, Smith, C. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1JBV
 
 | | FPGS-AMPPCP complex | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHASE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
8R1Q
 
 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) in complex with alpha-ketoamide 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Sun, X, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV-2 Omicron (P132H+T169S) complex with alpha-ketoamide 13b-K
To Be Published
|
|
1JBW
 
 | | FPGS-AMPPCP-folate complex | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, FOLYLPOLYGLUTAMATE SYNTHASE, ... | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
8R24
 
 | |
5YMU
 
 | |
5ZHG
 
 | |
5ZHO
 
 | |
5YMT
 
 | |
5YMS
 
 | |
4QN4
 
 | | Crystal structure of Neuraminidase N6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4QN6
 
 | | Crystal structure of Neuraminidase N6 complexed with Laninamivir | | Descriptor: | 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, Neuraminidase, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4QN7
 
 | | Crystal structure of neuramnidase N7 complexed with Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4QN5
 
 | | Neuraminidase N5 binds LSTa at the second SIA binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4QN3
 
 | | Crystal structure of Neuraminidase N7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
8IN4
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
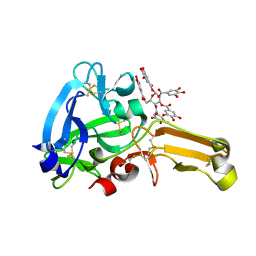 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
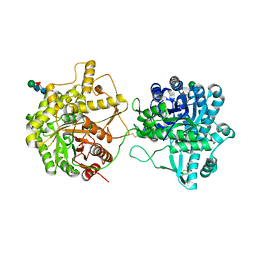 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
7XI4
 
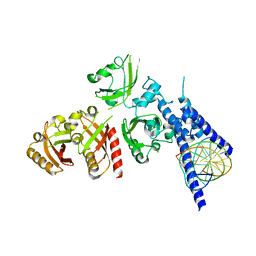 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.707 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
