8RNE
 
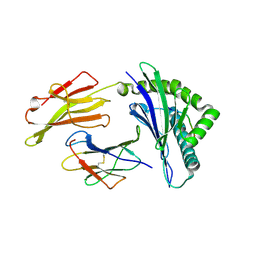 | | HLA-E*01:03 in complex with SARS-CoV-2 Nsp13 peptide, VMPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8RNF
 
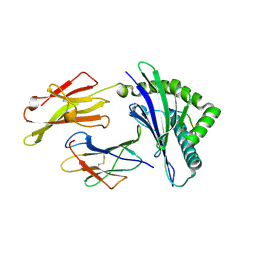 | | HLA-E*01:03 in complex with SARS-CoV-2 Omicron Nsp13 peptide, VIPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8XFD
 
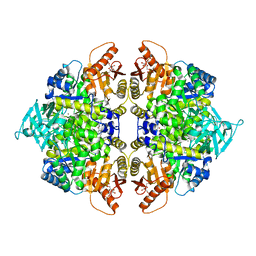 | | Crystal structure of pyruvate kinase tetramer in complex with allosteric activator, Mitapivat (MTPV, AG-348) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Isoform L-type of Pyruvate kinase PKLR, MAGNESIUM ION, ... | | Authors: | Sun, R, Achour, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Crystal Structure of the Pyruvate Kinase Tetramer in Complex with the Allosteric Activator Mitapivat/AG-348
Crystals, 2024
|
|
6FTX
 
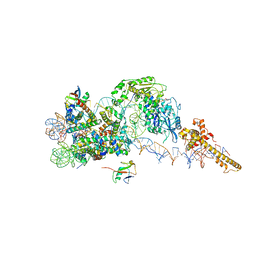 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
6G0L
 
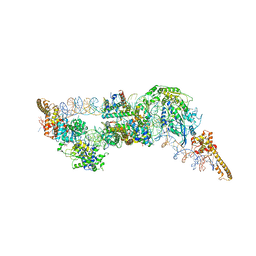 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
2AO5
 
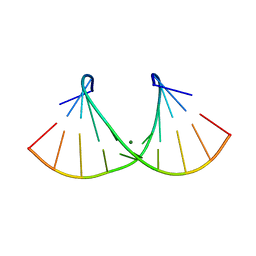 | | Crystal structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G-U base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*(5BU)P*GP*CP*GP*CP*U)-3', MAGNESIUM ION | | Authors: | Utsunomiya, R, Suto, K, Balasundaresan, D, Fukamizu, A, Kumar, P.K, Mizuno, H. | | Deposit date: | 2005-08-12 | | Release date: | 2006-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7EP8
 
 | |
7WU9
 
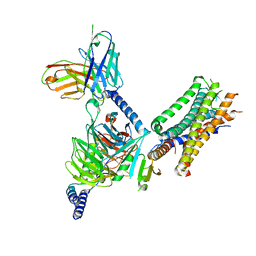 | | Cryo-EM structure of the human EP3-Gi signaling complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.375 Å) | | Cite: | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
6ICE
 
 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
7BUP
 
 | |
3SBJ
 
 | | MutM slanted complex 7 | | Descriptor: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*G)-3', 5'-D(P*CP*CP*TP*GP*GP*TP*(CX)P*TP*AP*CP*C)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Verdine, G.L. | | Deposit date: | 2011-06-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
3W91
 
 | | crystal structure of SeMet-labeled yeast N-acetyltransferase Mpr1 L87M mutant | | Descriptor: | MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-03-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
4G4N
 
 | | MutM containing M77A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
2C7Z
 
 | | Plant enzyme crystal form II | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
2C7Y
 
 | | plant enzyme | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Germain, V, Bryce, J.H, Smith, S.M, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
5ZKB
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
4G4Q
 
 | | MutM containing F114A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
4G4O
 
 | | MutM containing M77A mutation bound to oxoG-containing DNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
