6EV1
 
 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
5O15
 
 | | Crystal structure of bifunctional dehydratase-cyclase domain in ambruticin biosynthesis | | Descriptor: | AmbC, GLYCEROL | | Authors: | Sung, K.H, Berkhan, G, Hollmann, T, Wagner, L, Hahn, F, Blankenfeldt, W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Insights into the Dual Activity of a Bifunctional Dehydratase-Cyclase Domain.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5O16
 
 | | Crystal structure of bifunctional dehydratase-cyclase domain in ambruticin biosynthesis | | Descriptor: | AmbC, SULFATE ION | | Authors: | Sung, K.H, Berkhan, G, Hollmann, T, Wagner, L, Hahn, F, Blankenfeldt, W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Insights into the Dual Activity of a Bifunctional Dehydratase-Cyclase Domain.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5E1Q
 
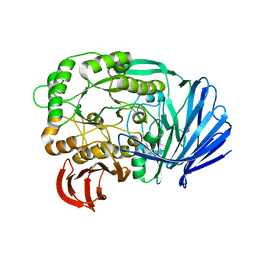 | | Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac | | Descriptor: | CALCIUM ION, GLYCEROL, Retaining alpha-galactosidase, ... | | Authors: | Matsunaga, K, Yamashita, K, Tagami, T, Yao, M, Okuyama, M, Kimura, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
FEBS J., 284, 2017
|
|
4HNZ
 
 | |
4HO7
 
 | |
3QKZ
 
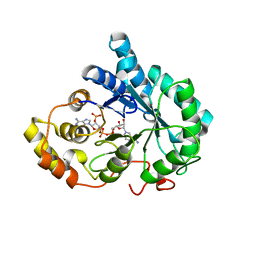 | | Crystal structure of mutant His269Arg AKR1B14 | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, El-Kabbani, O. | | Deposit date: | 2011-02-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of the His269Arg mutant of the rat aldose reductase-like protein AKR1B14 complexed with NADPH.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3O3R
 
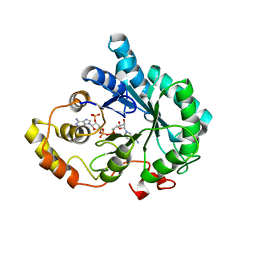 | | Crystal Structure of AKR1B14 in complex with NADP | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of rat aldose reductase-like protein AKR1B14 holoenzyme: Probing the role of His269 in coenzyme binding by site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5XQ1
 
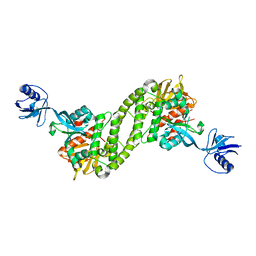 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPY
 
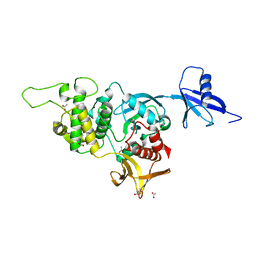 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
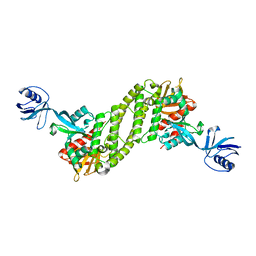 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6JOE
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase, ... | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2019-03-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
4UBQ
 
 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
1DTJ
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | RNA-BINDING NEUROONCOLOGICAL VENTRAL ANTIGEN 2 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L, Musunuru, K, Zhong, R, Darnell, R.B, Burley, S.K. | | Deposit date: | 2000-01-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1EC6
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN BOUND TO 20-MER RNA HAIRPIN | | Descriptor: | 20-MER RNA HAIRPIN, RNA-BINDING PROTEIN NOVA-2 | | Authors: | Lewis, H.A, Musunuru, K, Jensen, K.B, Edo, C, Chen, H. | | Deposit date: | 2000-01-25 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome.
Cell(Cambridge,Mass.), 100, 2000
|
|
7W7S
 
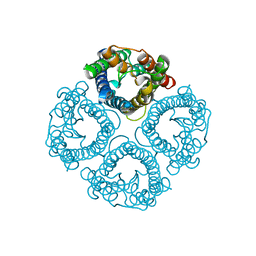 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7R
 
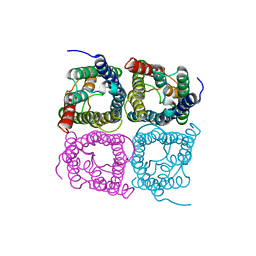 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
1HQS
 
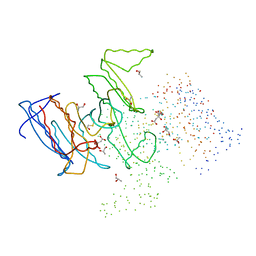 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
1K0R
 
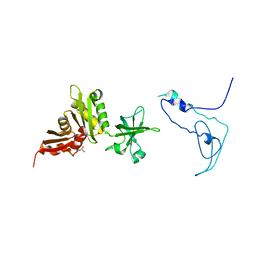 | | Crystal Structure of Mycobacterium tuberculosis NusA | | Descriptor: | NusA, SULFATE ION | | Authors: | Gopal, B, Haire, L.F, Gamblin, S.J, Dodson, E.J, Lane, A.N, Papavinasasundaram, K.G, Colston, M.J, Dodson, G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-09-20 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution.
J.Mol.Biol., 314, 2001
|
|
1C4L
 
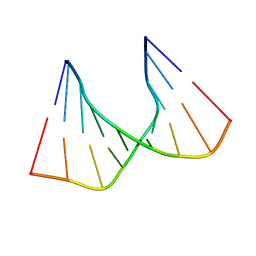 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
5ZHI
 
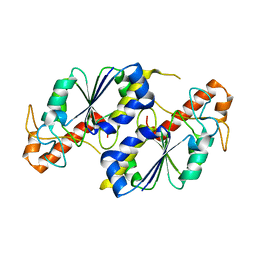 | | Apo crystal structure of TrmD from Mycobacterium tuberculosis | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5AX7
 
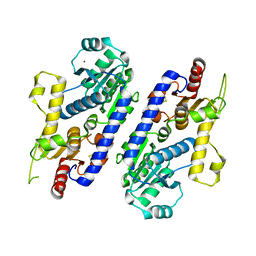 | | yeast pyruvyltransferase Pvg1p | | Descriptor: | Pyruvyl transferase 1, ZINC ION | | Authors: | Kanekiyo, M, Yoritsune, K, Yoshinaga, S, Higuchi, Y, Takegawa, K, Kakuta, Y. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-08 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A rationally engineered yeast pyruvyltransferase Pvg1p introduces sialylation-like properties in neo-human-type complex oligosaccharide
Sci Rep, 6, 2016
|
|
