7LHW
 
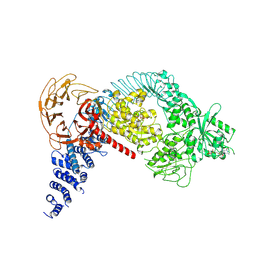 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
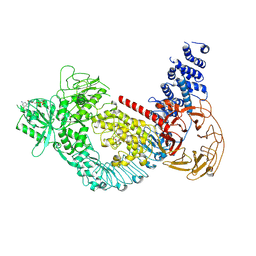 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI4
 
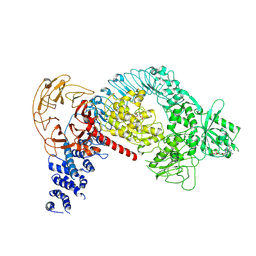 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
6V97
 
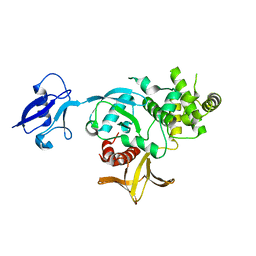 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
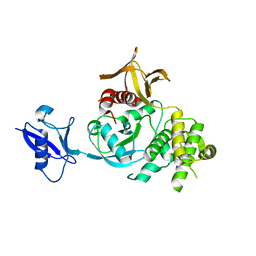 | | Kindlin-3 double deletion mutant long form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
1U5T
 
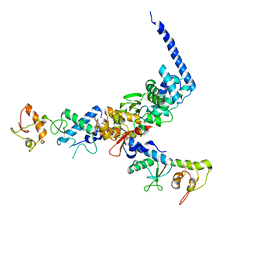 | | Structure of the ESCRT-II endosomal trafficking complex | | Descriptor: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | Authors: | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
5K5W
 
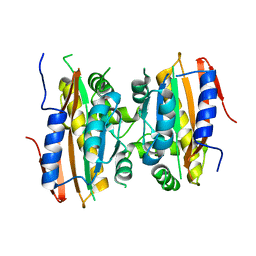 | | Crystal structure of limiting CO2-inducible protein LCIB | | Descriptor: | ZINC ION, limiting CO2-inducible protein LCIB | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3OWO
 
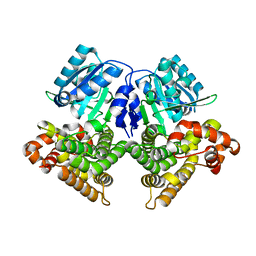 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
7RIC
 
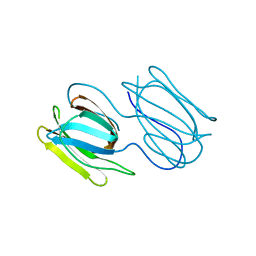 | | Griffithsin variant Y28W/Y68W/Y110W | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RID
 
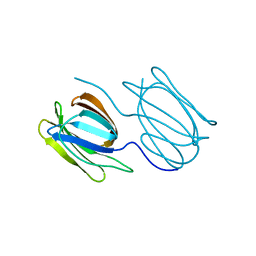 | | Griffithsin variant Y28A | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RKG
 
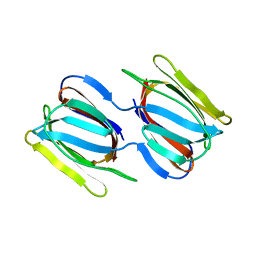 | | Griffithsin mutant Y28W | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
3OX4
 
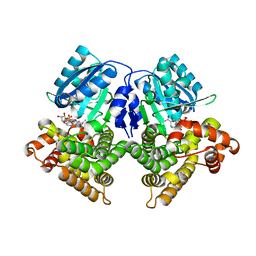 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
4AZN
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag-mediated crystal packing | | Descriptor: | FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZP
 
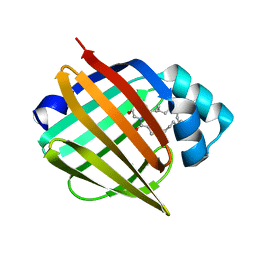 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZR
 
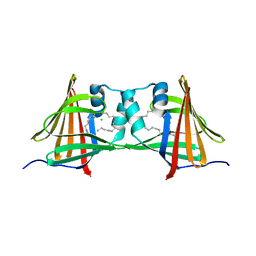 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
