1A0P
 
 | | SITE-SPECIFIC RECOMBINASE, XERD | | Descriptor: | SITE-SPECIFIC RECOMBINASE XERD | | Authors: | Subramanya, H.S, Arciszewska, L.K, Baker, R.A, Bird, L.E, Sherratt, D.J, Wigley, D.B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the site-specific recombinase, XerD.
EMBO J., 16, 1997
|
|
1A0I
 
 | | ATP-DEPENDENT DNA LIGASE FROM BACTERIOPHAGE T7 COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA LIGASE | | Authors: | Subramanya, H.S, Doherty, A.J, Ashford, S.R, Wigley, D.B. | | Deposit date: | 1997-12-01 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7.
Cell(Cambridge,Mass.), 85, 1996
|
|
1OTF
 
 | | 4-OXALOCROTONATE TAUTOMERASE-TRICLINIC CRYSTAL FORM | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1OTG
 
 | | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE | | Descriptor: | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE, SULFATE ION | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1PJR
 
 | |
4JX8
 
 | |
4JQC
 
 | | Crystal Structure of E.coli Enoyl Reductase in Complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Subramanya, H, Rao, K.N, Anirudha, L. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E.coli Enoyl Reductase in Complex with NAD and AFN-1252
TO BE PUBLISHED
|
|
4JZ1
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[(3-{[(4-fluorophenyl)sulfonyl]amino}pyridine-2,6-diyl)bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JZI
 
 | | Crystal Structure of Matriptase in complex with Inhibitor". | | Descriptor: | N-(trans-4-aminocyclohexyl)-2,6-bis(4-carbamimidoylphenoxy)pyridine-4-carboxamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Chandra, R.B, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JYT
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[{3-[(naphthalen-2-ylsulfonyl)amino]pyridine-2,6-diyl}bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4RLH
 
 | | Crystal structure of enoyl ACP reductase from Burkholderia pseudomallei in complex with AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Rao, K.N, Sarah, J, Anirudha, L, Subramanya, H.S. | | Deposit date: | 2014-10-17 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of enoyl ACP reductase from
Burkholderia pseudomallei in complex with AFN-1252
To be Published
|
|
2PJR
 
 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
3PJR
 
 | | HELICASE SUBSTRATE COMPLEX | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism
Cell(Cambridge,Mass.), 97, 1999
|
|
1YJS
 
 | | K226Q Mutant Of Serine Hydroxymethyltransferase From B. Stearothermophilus, Complex With Glycine | | Descriptor: | GLYCINE, PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Bhavani, S, Trivedi, V, Jala, V.R, Subramanya, H.S, Kaul, P, Purnima, K, Prakash, V, Appaji, R.N, Savithri, H.S. | | Deposit date: | 2005-01-15 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Lys-226 in the Catalytic Mechanism of Bacillus Stearothermophilus Serine Hydroxymethyltransferase-Crystal Structure and Kinetic Studies
Biochemistry, 44, 2005
|
|
1YJY
 
 | | K226M Mutant Of Serine Hydroxymethyltransferase From B. Stearothermophilus, Complex With Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Bhavani, S, Trivedi, V, Jala, V.R, Subramanya, H.S, Kaul, P, Purnima, K, Prakash, V, Appaji, R.N, Savithri, H.S. | | Deposit date: | 2005-01-16 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Lys-226 in the Catalytic Mechanism of Bacillus Stearothermophilus Serine Hydroxymethyltransferase-Crystal Structure and Kinetic Studies
Biochemistry, 44, 2005
|
|
1YJZ
 
 | | K226M Mutant Of Serine Hydroxymethyltransferase From B. Stearothermophilus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Bhavani, S, Trivedi, V, Jala, V.R, Subramanya, H.S, Kaul, P, Purnima, K, Prakash, V, Appaji, R.N, Savithri, H.S. | | Deposit date: | 2005-01-16 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Lys-226 in the Catalytic Mechanism of Bacillus Stearothermophilus Serine Hydroxymethyltransferase-Crystal Structure and Kinetic Studies
Biochemistry, 44, 2005
|
|
4R0I
 
 | | CRYSTAL STRUCTURE of MATRIPTASE in COMPLEX WITH INHIBITOR | | Descriptor: | 3-({(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-[(naphthalen-2-ylsulfonyl)amino]-3-oxopropyl}oxy)benzenecarboximidamide, SERINE PROTEASE, MATRIPTASE, ... | | Authors: | Rao, K.N, Ashok, K.N, Chakshusmathi, G, Rajeev, G, Subramanya, H. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1KL2
 
 | | Crystal Structure of Serine Hydroxymethyltransferase Complexed with Glycine and 5-formyl tetrahydrofolate | | Descriptor: | GLYCINE, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-11 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1I9G
 
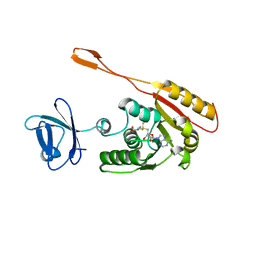 | | CRYSTAL STRUCTURE OF AN ADOMET DEPENDENT METHYLTRANSFERASE | | Descriptor: | HYPOTHETICAL PROTEIN RV2118C, S-ADENOSYLMETHIONINE | | Authors: | Gupta, A, Kumar, P.H, Dineshkumar, T.K, Varshney, U, Subramanya, H.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-03-20 | | Release date: | 2001-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv.
J.Mol.Biol., 312, 2001
|
|
1KKP
 
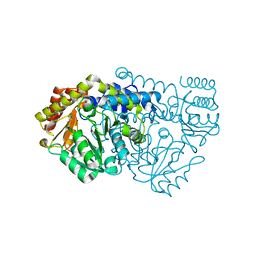 | | Crystal Structure of Serine Hydroxymethyltransferase complexed with Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KL1
 
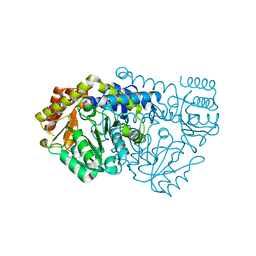 | | Crystal Structure of Serine Hydroxymethyltransferase Complexed with Glycine | | Descriptor: | GLYCINE, PYRIDOXAL-5'-PHOSPHATE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-11 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KKJ
 
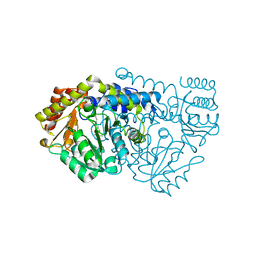 | | Crystal Structure of Serine Hydroxymethyltransferase from B.stearothermophilus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
4O9V
 
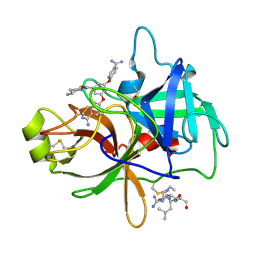 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis(4-carbamimidoylphenoxy)benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4O97
 
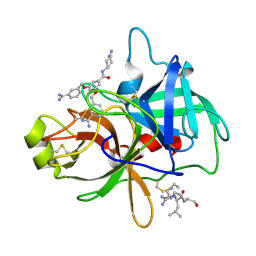 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4I8N
 
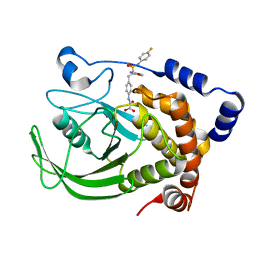 | | CRYSTAL STRUCTURE of PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN INHIBITOR [(4-{(2S)-2-(1,3-BENZOXAZOL-2-YL)-2-[(4-FLUOROPHENYL)SULFAMOYL]ETHYL}PHENYL)AMINO](OXO)ACETIC ACID | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, [(4-{(2S)-2-(1,3-benzoxazol-2-yl)-2-[(4-fluorophenyl)sulfamoyl]ethyl}phenyl)amino](oxo)acetic acid | | Authors: | Reddy, S.M.V.V.V, Rao, K.N, Subramanya, H. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of PTP1B in Complex with a New PTP1B Inhibitor.
Protein Pept.Lett., 21, 2014
|
|
