6O0Y
 
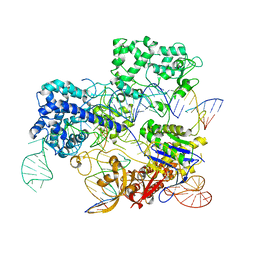 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3DNN
 
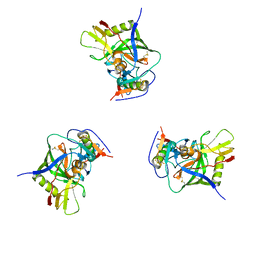 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6O0Z
 
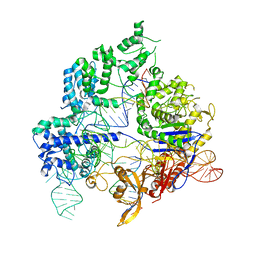 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, non-target strand DNA, single guide RNA, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3DNL
 
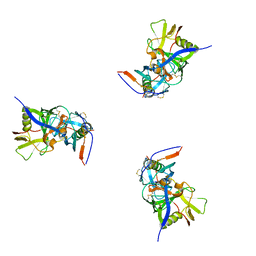 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6VYF
 
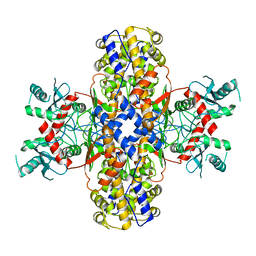 | | Cryo-EM structure of Plasmodium vivax hexokinase (Open state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6VYG
 
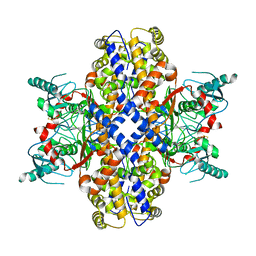 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
7RLG
 
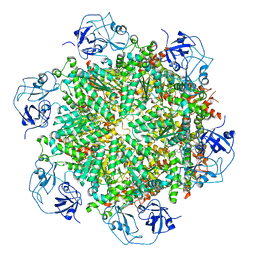 | | Cryo-EM structure of human p97-D592N mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RL9
 
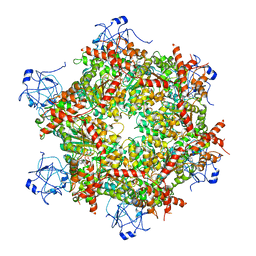 | | Cryo-EM structure of human p97-R191Q mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RL6
 
 | | Cryo-EM structure of human p97-R155H mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLB
 
 | | Cryo-EM structure of human p97-A232E mutant bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLD
 
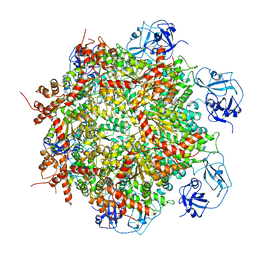 | | Cryo-EM structure of human p97-E470D mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
8DLK
 
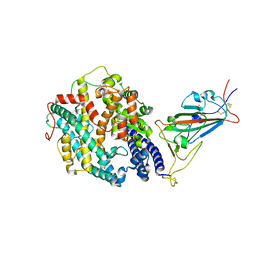 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLS
 
 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLV
 
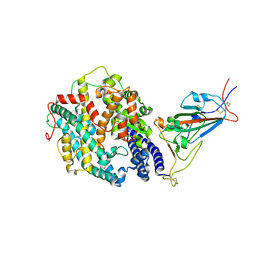 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
7RLI
 
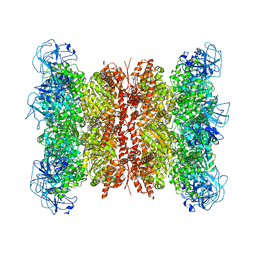 | | Cryo-EM structure of human p97 bound to CB-5083 and ADP. | | Descriptor: | 1-[4-(benzylamino)-7,8-dihydro-5H-pyrano[4,3-d]pyrimidin-2-yl]-2-methyl-1H-indole-4-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
8DLO
 
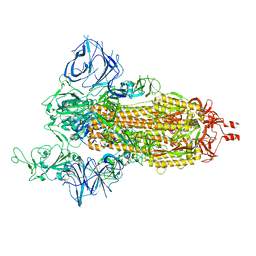 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLJ
 
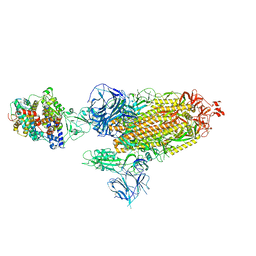 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLT
 
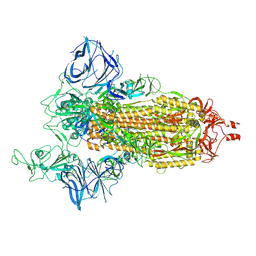 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLW
 
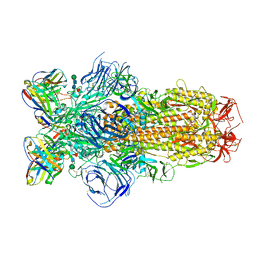 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with Fab S2M11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S2M11 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLY
 
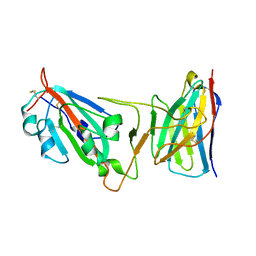 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 (focused refinement of NTD and VH ab6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab6 | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLU
 
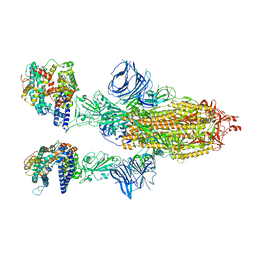 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLN
 
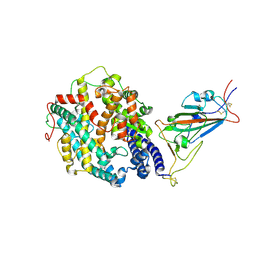 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLR
 
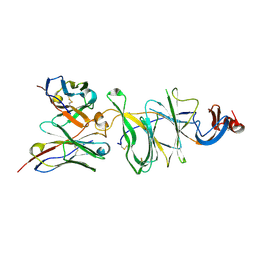 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4-8 (focused refinement of NTD and 4-8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4-8 heavy chain, Fab 4-8 light chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLP
 
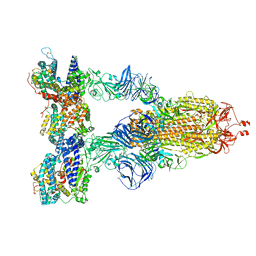 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLX
 
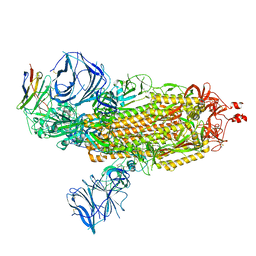 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
