3DFJ
 
 | | Crystal structure of human Prostasin | | Descriptor: | CHLORIDE ION, Prostasin | | Authors: | Su, H.P, Rickert, K.W, Darke, P.L, Munshi, S.K. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of human prostasin, a target for the regulation of hypertension.
J.Biol.Chem., 283, 2008
|
|
3DFL
 
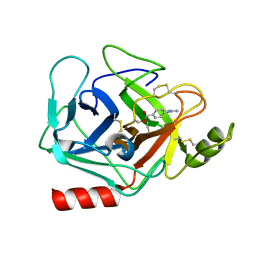 | | Crystal structure of human Prostasin complexed to 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, Prostasin | | Authors: | Su, H.P, Rickert, K.W, Darke, P.L, Munshi, S.K. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human prostasin, a target for the regulation of hypertension.
J.Biol.Chem., 283, 2008
|
|
3M9F
 
 | | HIV protease complexed with compound 10b | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-[(1S,5S)-5-{[(4-aminophenyl)sulfonyl](3-methylbutyl)amino}-1-methyl-6-oxohexyl]-Nalpha-(methoxycarbonyl)-beta-phenyl-L-phenylalaninamide | | Authors: | Su, H.P. | | Deposit date: | 2010-03-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsilon substituted lysinol derivatives as HIV-1 protease inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5I8A
 
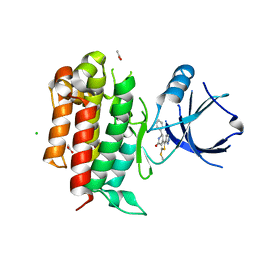 | | TrkA with (6~{R})-3-methylsulfanyl-6-phenyl-1-(1~{H}-pyrazol-3-yl)-6,7-dihydro-5~{H}-thieno[3,4-c]pyridin-4-one | | Descriptor: | (6R)-3-(methylsulfanyl)-6-phenyl-1-(1H-pyrazol-3-yl)-6,7-dihydrothieno[3,4-c]pyridin-4(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-02-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Potent, selective and orally bioavailable leucine-rich repeat kinase 2 (LRRK2) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
4PMS
 
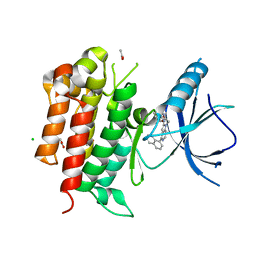 | | The structure of TrkA kinase bound to the inhibitor 4-naphthalen-1-yl-1-[(5-phenyl-1,2,4-oxadiazol-3-yl)methyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxylic acid | | Descriptor: | 4-(naphthalen-1-yl)-1-[(5-phenyl-1,2,4-oxadiazol-3-yl)methyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Maximizing diversity from a kinase screen: identification of novel and selective pan-Trk inhibitors for chronic pain.
J.Med.Chem., 57, 2014
|
|
4PMP
 
 | |
4PMM
 
 | |
4PMT
 
 | |
8T9Z
 
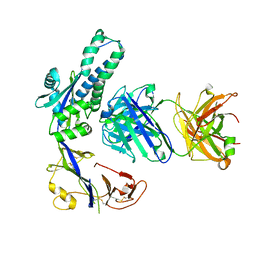 | | Structural of M8C10 Fab in complex human metapneumovirus fusion protein | | Descriptor: | Fusion glycoprotein F0, M8C10 Fab Heavy Chain, M8C10 Fab Light Chain | | Authors: | Su, H.P, Eddins, M.J, Shipman, J.M, Kostas, J, Reid, J.C. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural characterization of M8C10, a neutralizing antibody targeting a highly conserved prefusion-specific epitope on the metapneumovirus fusion trimerization interface.
J.Virol., 97, 2023
|
|
2G2Q
 
 | | The crystal structure of G4, the poxviral disulfide oxidoreductase essential for cytoplasmic disulfide bond formation | | Descriptor: | Glutaredoxin-2, SULFATE ION | | Authors: | Su, H.P, Lin, D.Y, Garboczi, D.N. | | Deposit date: | 2006-02-16 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of G4, the poxvirus disulfide oxidoreductase essential for virus maturation and infectivity.
J.Virol., 80, 2006
|
|
2I9L
 
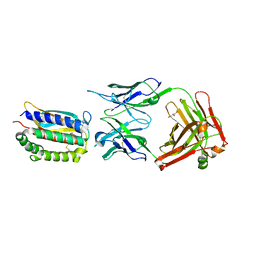 | | Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein | | Descriptor: | Antibody 7D11 heavy chain, Antibody 7D11 light chain, GLYCEROL, ... | | Authors: | Su, H.P, Golden, J.W, Gittis, A.G, Moss, B, Hooper, J.W, Garboczi, D.N. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Virology, 368, 2007
|
|
6OUS
 
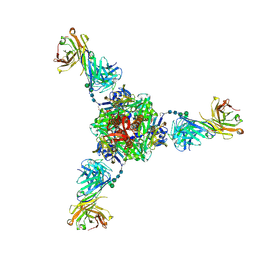 | | Structure of fusion glycoprotein from human respiratory syncytial virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Su, H.P. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein.
Nat Commun, 10, 2019
|
|
6XDS
 
 | |
1YPY
 
 | | Crystal Structure of Vaccinia Virus L1 protein | | Descriptor: | Virion membrane protein | | Authors: | Su, H.P, Garman, S.C, Allison, T.J, Fogg, C, Moss, B, Garboczi, D.N. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The 1.51-Angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5HX8
 
 | |
6B3F
 
 | |
6B38
 
 | |
6B3G
 
 | |
5IVT
 
 | | Crystal Structure of HIV Protease complexed with [(1S)-1-[(S)-(4-chlorophenyl)-(3,5-difluorophenyl)methyl]-2-[[5-fluoro-4-[2-[(2R,5S)-5-(2,2,2-trifluoroethylcarbamoyloxymethyl)morpholin-4-ium-2-yl]ethyl]pyridin-1-ium-3-yl]amino]-2-oxo-ethyl]ammonium | | Descriptor: | (betaS)-4-chloro-beta-(3,5-difluorophenyl)-N-(5-fluoro-4-{2-[(2R,5S)-5-({[(2,2,2-trifluoroethyl)carbamoyl]oxy}methyl)morpholin-2-yl]ethyl}pyridin-3-yl)-L-phenylalaninamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-03-21 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of MK-8718, an HIV Protease Inhibitor Containing a Novel Morpholine Aspartate Binding Group.
Acs Med.Chem.Lett., 7, 2016
|
|
5IVR
 
 | |
5IVQ
 
 | |
6B36
 
 | |
6B3C
 
 | |
6B3H
 
 | | Crystal Structure of HIV Protease complexed with N-(2-(2-((6R,9S)-2,2-dioxido-2-thia-1,7-diazabicyclo[4.3.1]decan-9-yl)ethyl)-3-fluorophenyl)-3,3-bis(4-fluorophenyl)propanamide | | Descriptor: | CHLORIDE ION, HIV-1 Protease, N-(2-{2-[(6R,9S)-2,2-dioxo-2lambda~6~-thia-1,7-diazabicyclo[4.3.1]decan-9-yl]ethyl}-3-fluorophenyl)-3,3-bis(4-fluorophenyl)propanamide | | Authors: | Su, H.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design and Synthesis of Piperazine Sulfonamide Cores Leading to Highly Potent HIV-1 Protease Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5KML
 
 | |
