2WQP
 
 | | Crystal structure of sialic acid synthase NeuB-inhibitor complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, 5-(ACETYLAMINO)-3,5-DIDEOXY-2-O-PHOSPHONO-D-ERYTHRO-L-MANNO-NONONIC ACID, ... | | Authors: | Liu, F, Lee, H.J, Strynadka, N.C.J, Tanner, M.E. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Inhibition of Neisseria Meningitidis Sialic Acid Synthase by a Tetrahedral Intermediate Analog.
Biochemistry, 48, 2009
|
|
2WNB
 
 | | Crystal Structure of a Mammalian Sialyltransferase in complex with disaccharide and CMP | | Descriptor: | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 1, CYTIDINE-5'-MONOPHOSPHATE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, ... | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WLD
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSIALIC ACID O-ACETYLTRANSFERASE | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
2X63
 
 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2X62
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2WLF
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
2WLE
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COENZYME A, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
2WLG
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSIALIC ACID O-ACETYLTRANSFERASE, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
2X49
 
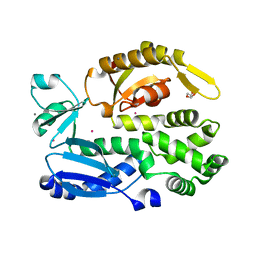 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
7KGM
 
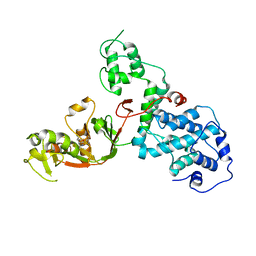 | | C. rodentium YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Putative exported protein | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-17 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
7LQ6
 
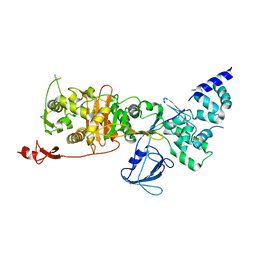 | | CryoEM structure of Escherichia coli PBP1b | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
7JOY
 
 | |
7K7K
 
 | |
7JP1
 
 | |
7K08
 
 | |
7LBU
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase | | Descriptor: | ACETATE ION, Exo-alpha-sialidase, PHOSPHATE ION | | Authors: | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
7LBV
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Exo-alpha-sialidase, PHOSPHATE ION | | Authors: | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | Deposit date: | 2021-01-09 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
7KCX
 
 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCV
 
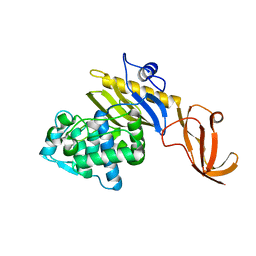 | |
7KCY
 
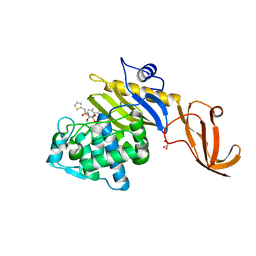 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7JLR
 
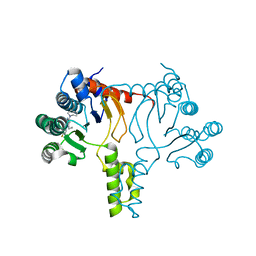 | |
7JLM
 
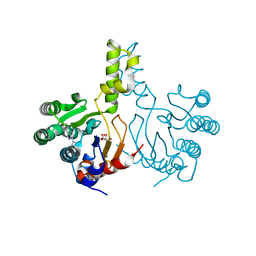 | |
7JLJ
 
 | |
7JLI
 
 | | Crystal structure of Bacillus subtilis UppS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Isoprenyl transferase | | Authors: | Workman, S.D, Strynadka, N.C.J. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Inhibition of Undecaprenyl Pyrophosphate Synthase from Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
7KGN
 
 | | S. Typhi YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, L,D-transpeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
