1AVS
 
 | |
3L7J
 
 | |
3L7K
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | 著者 | Strynadka, N.C.J, Lovering, A.L. | | 登録日 | 2009-12-28 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7I
 
 | |
6DCS
 
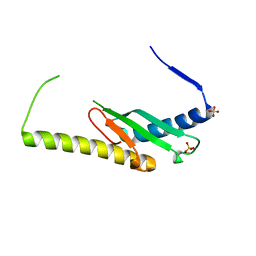 | | Stage III sporulation protein AF (SpoIIIAF) | | 分子名称: | SULFATE ION, Stage III sporulation protein AF | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2018-05-08 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6BS9
 
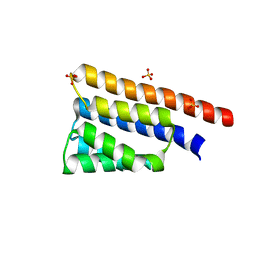 | | Stage III sporulation protein AB (SpoIIIAB) | | 分子名称: | SULFATE ION, Stage III sporulation protein AB | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2017-12-01 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural characterization of SpoIIIAB sporulation-essential protein in Bacillus subtilis.
J. Struct. Biol., 202, 2018
|
|
3SPU
 
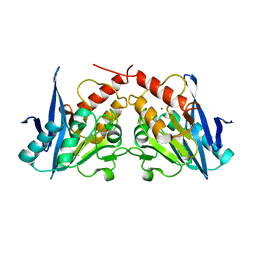 | | apo NDM-1 Crystal Structure | | 分子名称: | Beta-lactamase NDM-1, ZINC ION | | 著者 | Strynadka, N.C.J, King, D.T. | | 登録日 | 2011-07-03 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance
Protein Sci., 20, 2011
|
|
1PPK
 
 | |
1JTG
 
 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | 分子名称: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | 著者 | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | 登録日 | 2001-08-20 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
4EY2
 
 | | Crystal structure of NDM-1 bound to hydrolyzed methicillin | | 分子名称: | (2R,4S)-2-{(R)-carboxy[(2,6-dimethoxybenzoyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | 著者 | Strynadka, N.C.J, King, D.T. | | 登録日 | 2012-05-01 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EXY
 
 | |
4EYF
 
 | | Crystal structure of NDM-1 bound to hydrolyzed benzylpenicillin | | 分子名称: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | 著者 | Strynadka, N.C.J, King, D.T. | | 登録日 | 2012-05-01 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EXS
 
 | |
4EYL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem | | 分子名称: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | 著者 | Strynadka, N.C.J, King, D.T. | | 登録日 | 2012-05-01 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EYB
 
 | | Crystal structure of NDM-1 bound to hydrolyzed oxacillin | | 分子名称: | (2R,4S)-2-[(R)-carboxy{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carbo xylic acid, Beta-lactamase NDM-1, ZINC ION | | 著者 | Strynadka, N.C.J, King, D.T. | | 登録日 | 2012-05-01 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | 分子名称: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | 著者 | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | 登録日 | 2009-03-15 | | 公開日 | 2009-03-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
8SXR
 
 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | 分子名称: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | 登録日 | 2023-05-23 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
6ZTG
 
 | | Spor protein DedD | | 分子名称: | Cell division protein DedD | | 著者 | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | 登録日 | 2020-07-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | 分子名称: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | 著者 | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | 登録日 | 2020-03-13 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
7LBV
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase in complex with Neu5Ac2en | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Exo-alpha-sialidase, PHOSPHATE ION | | 著者 | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | 登録日 | 2021-01-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
7LBU
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase | | 分子名称: | ACETATE ION, Exo-alpha-sialidase, PHOSPHATE ION | | 著者 | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | 登録日 | 2021-01-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | 著者 | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | 登録日 | 2017-07-04 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
5WCN
 
 | |
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
