3BZZ
 
 | | Crystal structural of the mutated R313T EscU/SpaS C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.407 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
6PEE
 
 | |
3BZO
 
 | | Crystal structural of native EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZV
 
 | | Crystal structural of the mutated T264A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
6PEM
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgHK from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q14
 
 | | Structure of the Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
3UJZ
 
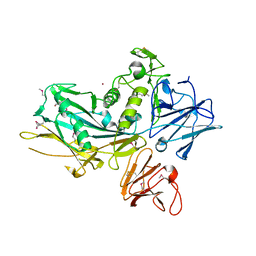 | |
4G2S
 
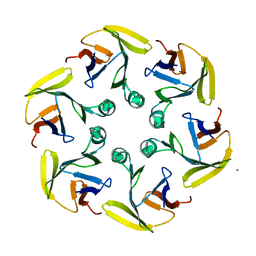 | |
4G1I
 
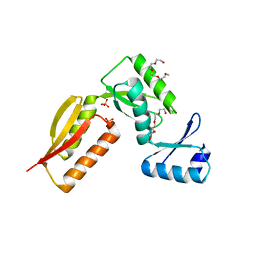 | |
4DKI
 
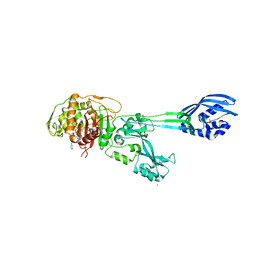 | | Structural Insights into the Anti- Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BICARBONATE ION, CADMIUM ION, ... | | Authors: | Lovering, A.L, Gretes, M.C, Strynadka, N.C.J. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Anti-methicillin-resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole.
J.Biol.Chem., 287, 2012
|
|
6N7O
 
 | | Crystal structure of GIL01 gp7 | | Descriptor: | GIL01 gp7, IODIDE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure, 27, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NTZ
 
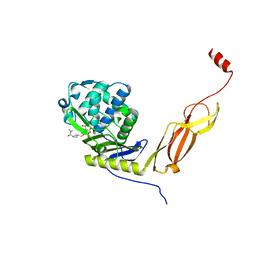 | | Crystal structure of E. coli PBP5-meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6NTW
 
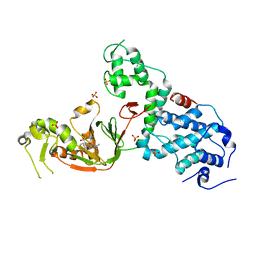 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6O9S
 
 | |
6NJP
 
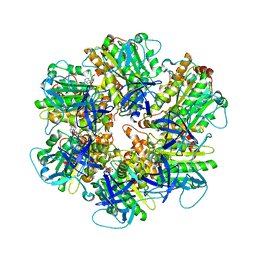 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6O9W
 
 | |
4G08
 
 | |
1VQQ
 
 | |
2QF5
 
 | |
5FA7
 
 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
2QF4
 
 | |
