7KCY
 
 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7JP1
 
 | |
7JOY
 
 | |
7KHP
 
 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
7LQ6
 
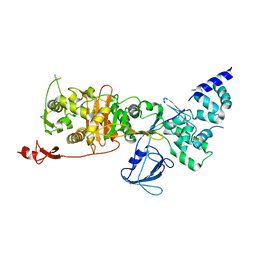 | | CryoEM structure of Escherichia coli PBP1b | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
2OSX
 
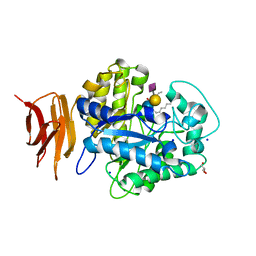 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | Descriptor: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
5TZ8
 
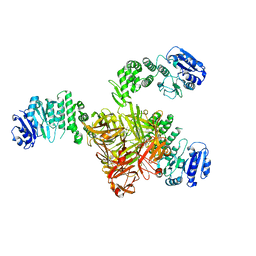 | | Crystal structure of S. aureus TarS | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TW4
 
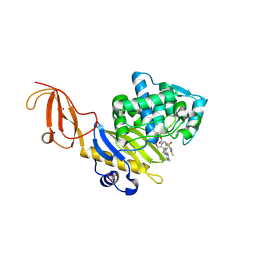 | |
5TZK
 
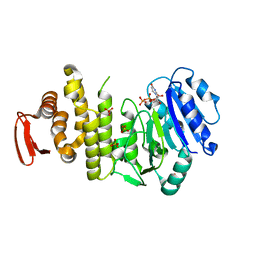 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TXI
 
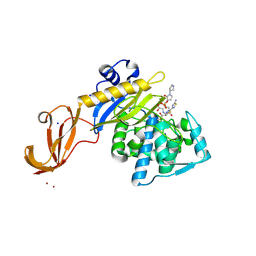 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
6XHS
 
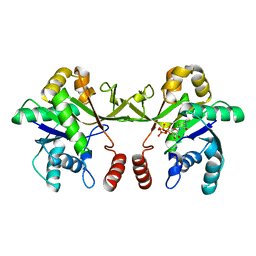 | | Crystal structure of S. aureus TarI in complex with CTP (space group P1211) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribitol-5-phosphate cytidylyltransferase 1, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of TarI and TarJ, a cytidylyltransferase and reductase pair for CDP-ribitol synthesis in Staphylococcus aureus wall teichoic acid biogenesis.
J.Struct.Biol., 213, 2021
|
|
6XHR
 
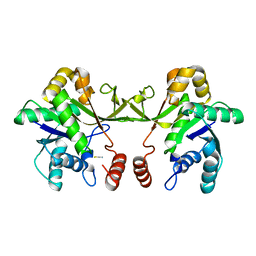 | | Crystal structure of S. aureus TarI (space group P1211) | | Descriptor: | Ribitol-5-phosphate cytidylyltransferase 1, THIOCYANATE ION | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis of TarI and TarJ, a cytidylyltransferase and reductase pair for CDP-ribitol synthesis in Staphylococcus aureus wall teichoic acid biogenesis.
J.Struct.Biol., 213, 2021
|
|
5TZI
 
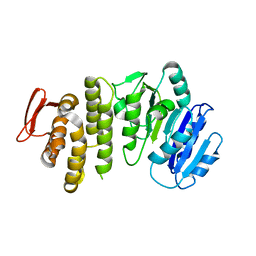 | | Crystal structure of S. aureus TarS 1-349 | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5U02
 
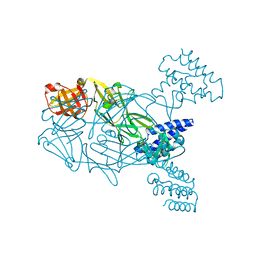 | | Crystal structure of S. aureus TarS 217-571 | | Descriptor: | Glycosyl transferase, IMIDAZOLE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-22 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
2X63
 
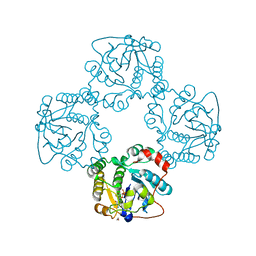 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2ZI8
 
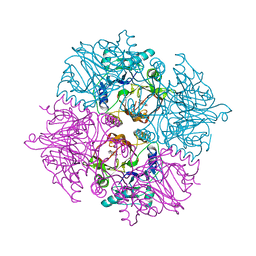 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
2X62
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
5TY2
 
 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TZJ
 
 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
3AFF
 
 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
5TY7
 
 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TZE
 
 | | Crystal structure of S. aureus TarS in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
2X49
 
 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
5TX9
 
 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TW8
 
 | |
