4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
4FMW
 
 | | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2 | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RNA (guanine-9-)-methyltransferase domain-containing protein 2, ... | | Authors: | Dong, A, Zeng, H, Loppnau, P, Tempel, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2
TO BE PUBLISHED
|
|
7QGP
 
 | | STK10 (LOK) bound to Macrocycle CKJB51 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-chloranyl-2-methyl-phenyl)methyl]-3-(7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaen-5-yl)urea, CHLORIDE ION, ... | | Authors: | Berger, B.-T, Schroeder, M, Kraemer, A, Schwalm, M.P, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STK10 bound to CKJB51
To Be Published
|
|
9GLQ
 
 | | Crystal structure of p73 tetramerisation domain in complex with darpins 1800 | | Descriptor: | COBALT (II) ION, Darpins 1800, GLYCEROL, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins as a novel tool to detect and degrade p73.
Cell Death Dis, 15, 2024
|
|
8P9E
 
 | | Crystal structure of wild type p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | Darpin 1810 F11, GLYCEROL, Isoform 2 of Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8T55
 
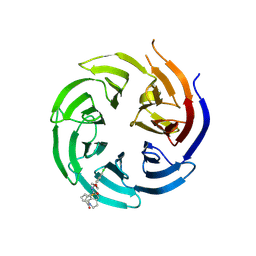 | | Co-crystal structure of the WD-repeat domain of human WDR91 in complex with MR46654 | | Descriptor: | 1,2-ETHANEDIOL, N-[3-(4-chlorophenyl)oxetan-3-yl]-1-propanoyl-1,2,3,4-tetrahydroquinoline-5-carboxamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Yen, H, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Santhakumar, V, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
8SHJ
 
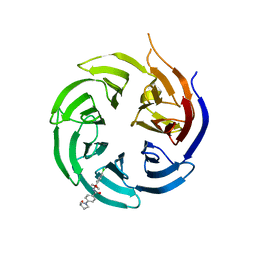 | | Crystal structure of the WD-repeat domain of human WDR91 in complex with MR45279 | | Descriptor: | N-[3-(4-chlorophenyl)oxetan-3-yl]-4-[(3S)-3-hydroxypyrrolidin-1-yl]benzamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
8SWJ
 
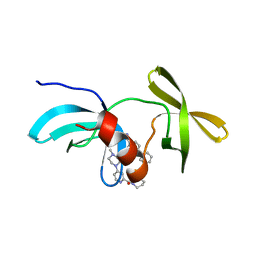 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531 | | Descriptor: | (3S)-N-(4'-carbamoyl[1,1'-biphenyl]-3-yl)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Pearce, K.H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531
To be published
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7YXY
 
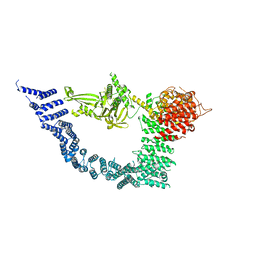 | |
7YXX
 
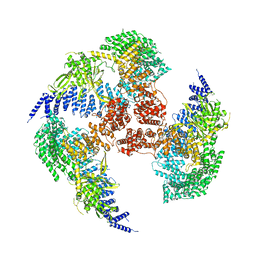 | |
9GII
 
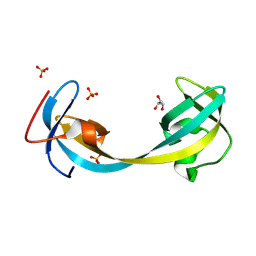 | | Jumonji domain-containing protein 2A with crystallization epitope mutation R913A | | Descriptor: | GLYCEROL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9GLE
 
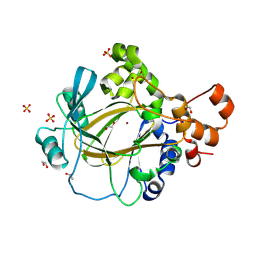 | | Jumonji domain-containing protein 2A with crystallization epitope mutations A91T:T93S | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
3GRO
 
 | | Human palmitoyl-protein thioesterase 1 | | Descriptor: | Palmitoyl-protein thioesterase 1, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Seitova, A, Tong, Y, Tempel, W, Dong, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Cossar, D, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Human palmitoyl-protein thioesterase 1
To be Published
|
|
9GP4
 
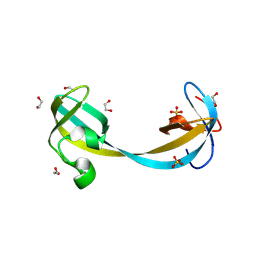 | | Jumonji domain-containing protein 2A with crystallization epitope mutations Q953E:A958D | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9GP1
 
 | | Jumonji domain-containing protein 2A with crystallization epitope mutatios K330R:A334E | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9H44
 
 | | Jumonji domain-containing protein 2B with crown ether and crystallization epitope mutations L916G:R917A:A918D | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Lysine-specific demethylase 4B | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-10-17 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
8R5D
 
 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
8QZ4
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb76) | | Descriptor: | BARIUM ION, CHOLESTEROL HEMISUCCINATE, Nanobody 76, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
7Q2J
 
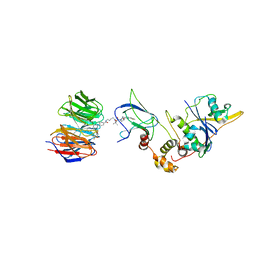 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8QZ2
 
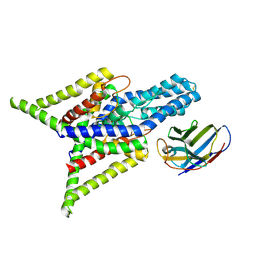 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an inhibitory nanobody (Nb61) | | Descriptor: | Nanobody 61, POTASSIUM ION, Potassium channel subfamily K member 10 | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ3
 
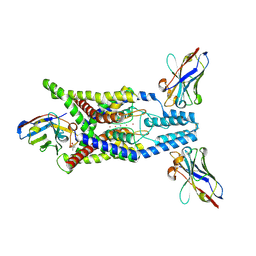 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
6V8W
 
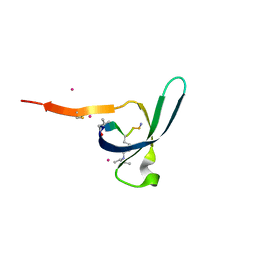 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
