5KTJ
 
 | | Crystal structure of Pistol, a class of self-cleaving ribozyme | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, Pistol (50-MER), ... | | Authors: | Nguyen, L.A, Wang, J, Steitz, T.A. | | Deposit date: | 2016-07-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of Pistol, a class of self-cleaving ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5J4B
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (co-crystallized) and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
1REA
 
 | |
1TV6
 
 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1WAF
 
 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1WAJ
 
 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1QVG
 
 | | Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1Q86
 
 | | Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q7Y
 
 | | Crystal Structure of CCdAP-Puromycin bound at the Peptidyl transferase center of the 50S ribosomal subunit | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q81
 
 | | Crystal Structure of minihelix with 3' puromycin bound to A-site of the 50S ribosomal subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2PYL
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(CTGACGAATGTACA)-3', 5'-d(GACTGCTTAC(2DA))-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYJ
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-d(ACACGTAAGCAGTC)-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2Q6T
 
 | |
2Q7E
 
 | | The structure of pyrrolysyl-tRNA synthetase bound to an ATP analogue | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q7G
 
 | | Pyrrolysine tRNA Synthetase bound to a pyrrolysine analogue (cyc) and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QLN
 
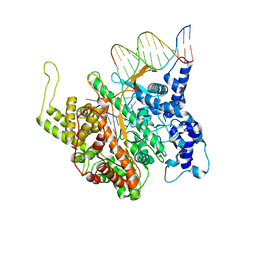 | | STRUCTURE OF A TRANSCRIBING T7 RNA POLYMERASE INITIATION COMPLEX | | Descriptor: | BACTERIOPHAGE T7 RNA POLYMERASE, DNA (5- D (P*CP*TP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP* AP*TP*TP*A)-3), DNA (5-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3), ... | | Authors: | Cheetham, G.M.T, Steitz, T.A. | | Deposit date: | 1999-09-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Transcribing T7 RNA Polymerase Initiation Complex
Science, 286, 1999
|
|
4V9R
 
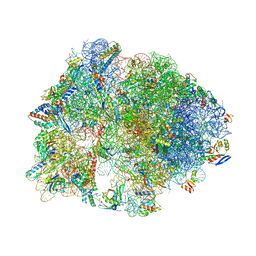 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V7Y
 
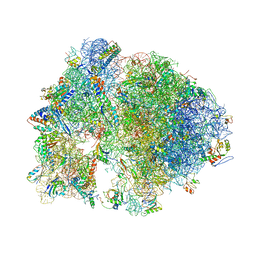 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
4V7X
 
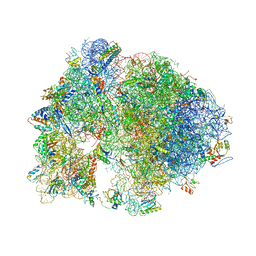 | | Structure of the Thermus thermophilus ribosome complexed with erythromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
354D
 
 | | Structure of loop E FROM E. coli 5S RRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*G)-3'), RNA-DNA (5'-R(*GP*CP*GP*AP*GP*AP*GP*UP*AP*)-D(*DGP(S)*)-R(*GP*C)-3') | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-10-01 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
357D
 
 | | 3.5 A structure of fragment I from E. coli 5S RRNA | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, RNA (5'-R(*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*G P*GP*AP*AP*CP*UP* GP*CP*CP*AP*GP*GP*CP*AP*U)-3'), ... | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-10-09 | | Release date: | 1997-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
