4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
8GJV
 
 | |
1T6V
 
 | | Crystal structure analysis of the nurse shark new antigen receptor (NAR) variable domain in complex with lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, novel antigen receptor | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
2I26
 
 | |
2I25
 
 | |
5E99
 
 | | Bovine Fab fragment F08_B11 | | Descriptor: | Fab F08_B11 heavy chain, Fab F08_B11 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
3OZ9
 
 | |
3Q1S
 
 | | HIV-1 neutralizing antibody Z13e1 in complex with epitope display protein | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Interleukin-22, ... | | Authors: | Stanfield, R.L, Julien, J.-P, Pejchal, R, Gach, J.S, Zwick, M.B, Wilson, I.A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Protein Immunogen that Displays an HIV-1 gp41 Neutralizing Epitope.
J.Mol.Biol., 414, 2011
|
|
3F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120 (MN ISOLATE); H315S MUTATION | | Descriptor: | PROTEIN (CYCLIC PEPTIDE (GP120)), PROTEIN (IMMUNOGLOBULIN GAMMA I (58.2)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-23 | | Release date: | 1999-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
5ILT
 
 | | Crystal structure of bovine Fab A01 | | Descriptor: | bovine Fab A01 heavy chain, bovine Fab A01 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5J74
 
 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
5J75
 
 | | Fluorogen Activating Protein AM2.2 in complex with ML342 | | Descriptor: | N,4-dimethyl-N-{2-oxo-2-[4-(pyridin-2-yl)piperazin-1-yl]ethyl}benzene-1-sulfonamide, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
5IJV
 
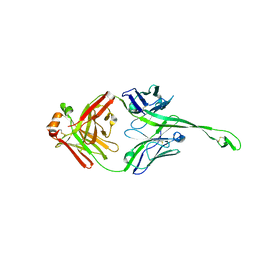 | | Crystal structure of bovine Fab E03 | | Descriptor: | bovine Fab E03 heavy chain, bovine Fab E03 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
2B1A
 
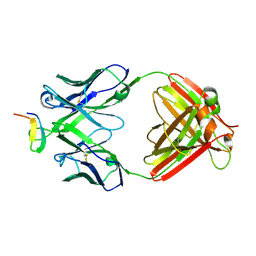 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2B1H
 
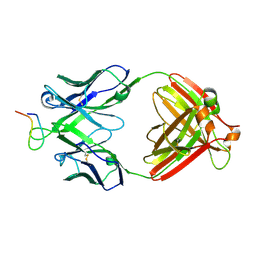 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG29 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2B0S
 
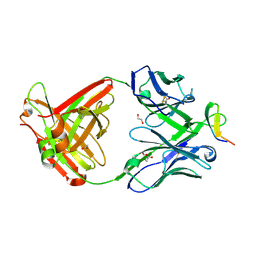 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with MN peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Fab 2219, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-14 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2OK0
 
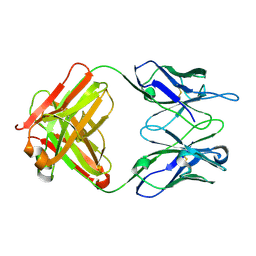 | | Fab ED10-DNA complex | | Descriptor: | 5'-D(*TP*C)-3', Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
6CXL
 
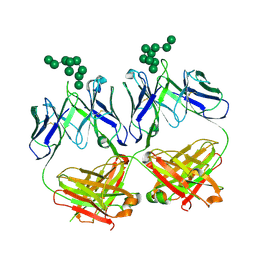 | | anti-HIV-1 Fab 2G12 in complex with glycopeptide 10F5 | | Descriptor: | alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose, anti-HIV-1 Fab 2G12 heavy chain, anti-HIV-1 Fab 2G12 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-04-03 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.586 Å) | | Cite: | Oligomannose Glycopeptide Conjugates Elicit Antibodies Targeting the Glycan Core Rather than Its Extremities.
ACS Cent Sci, 5, 2019
|
|
6OPA
 
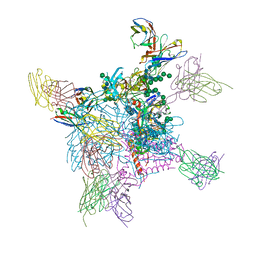 | |
6OO0
 
 | | Crystal structure of bovine Fab NC-Cow1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NC-Cow1 heavy chain, NC-Cow1 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
6CXG
 
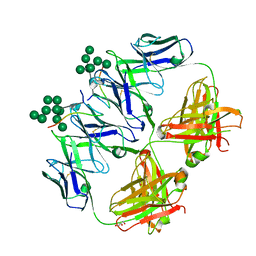 | | anti-HIV-1 Fab 2G12 in complex with glycopeptide 10V1S | | Descriptor: | 10V1S glycopeptide, GLYCEROL, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-04-03 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Oligomannose Glycopeptide Conjugates Elicit Antibodies Targeting the Glycan Core Rather than Its Extremities.
ACS Cent Sci, 5, 2019
|
|
5T3S
 
 | | HIV gp140 trimer MD39-10MUTA in complex with Fabs PGT124 and 35022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies.
Immunity, 45, 2016
|
|
5U65
 
 | | Camel Nanobody VHH-5 | | Descriptor: | SULFATE ION, VHH-5 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
5U64
 
 | | Camel nanobody VHH-28 | | Descriptor: | VHH-28 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
8EDF
 
 | |
