4XNQ
 
 | | Antibody hemagglutinin Complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, H5.3 Light chain, H5.3 heavy chain, ... | | Authors: | Spiller, B.W, Winarski, K.L. | | Deposit date: | 2015-01-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XRC
 
 | | Antibody hemagglutinin Complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, H5.3 Fab heavy chain, H5.3 Fab light chain, ... | | Authors: | Spiller, B.W, Winarski, K.L. | | Deposit date: | 2015-01-20 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3P2D
 
 | |
3QC1
 
 | | Protein Phosphatase Subunit: Alpha4 | | Descriptor: | Immunoglobulin-binding protein 1 | | Authors: | Spiller, B.W, LeNoue-Newton, M.L, Watkins, G.R, Germane, K.L, Zhou, P, McCorvey, L.R, Wadzinski, B.E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Mid1 and PP2Ac binding domains of Alpha4 are both required for Alpha4 to inhibit PP2Ac degradation
TO BE PUBLISHED
|
|
3Q1C
 
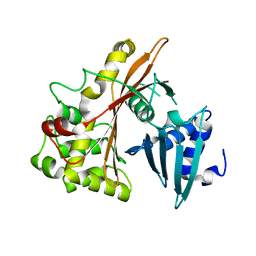 | | Structure of EspG Protein | | Descriptor: | LEE-encoded effector EspG | | Authors: | Spiller, B.W, Germane, K.L. | | Deposit date: | 2010-12-17 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural and Functional Studies Indicate That the EPEC Effector, EspG, Directly Binds p21-Activated Kinase.
Biochemistry, 50, 2011
|
|
4ELD
 
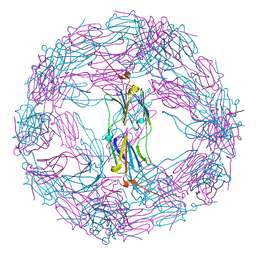 | |
4HFW
 
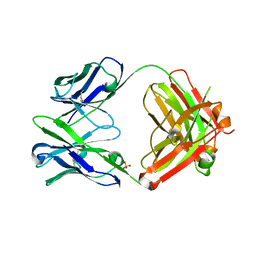 | | Anti Rotavirus Antibody | | Descriptor: | 6-26 Fab Heavy chain, 6-26 Fab Light chain, SULFATE ION | | Authors: | Spiller, B.W, Aiyegbo, M, Crowe, J.E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Human Rotavirus VP6-Specific Antibodies Mediate Intracellular Neutralization by Binding to a Quaternary Structure in the Transcriptional Pore.
Plos One, 8, 2013
|
|
4GSD
 
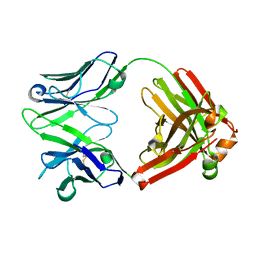 | | H5.3 Fab Structure | | Descriptor: | H5.3 Fab Heavy Chain, H5.3 Fab Light Chain | | Authors: | Spiller, B.W, Winarski, K.L. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses.
J.Clin.Invest., 123, 2013
|
|
1A4J
 
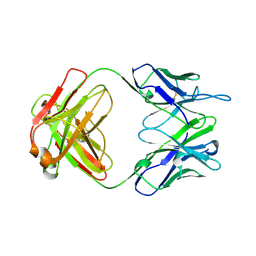 | | DIELS ALDER CATALYTIC ANTIBODY GERMLINE PRECURSOR | | Descriptor: | IMMUNOGLOBULIN, DIELS ALDER CATALYTIC ANTIBODY (HEAVY CHAIN), DIELS ALDER CATALYTIC ANTIBODY (LIGHT CHAIN) | | Authors: | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
1A4K
 
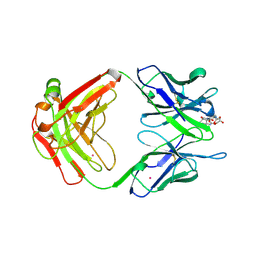 | | DIELS ALDER CATALYTIC ANTIBODY WITH TRANSITION STATE ANALOGUE | | Descriptor: | ANTIBODY FAB, CADMIUM ION, [4-(4-ACETYLAMINO-PHENYL)-3,5-DIOXO-4-AZA-TRICYCLO[5.2.2.0 2,6]UNDEC-1-YLCARBAMOYLOXY]-ACETIC ACID | | Authors: | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
4NRH
 
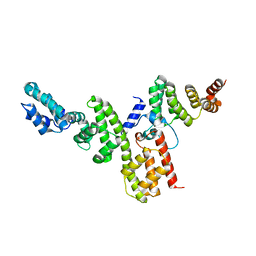 | | CopN-Scc3 complex | | Descriptor: | Chaperone SycD, CopN, SODIUM ION | | Authors: | Archuleta, T.L, Spiller, B.W. | | Deposit date: | 2013-11-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A gatekeeper chaperone complex directs translocator secretion during type three secretion.
Plos Pathog., 10, 2014
|
|
4R04
 
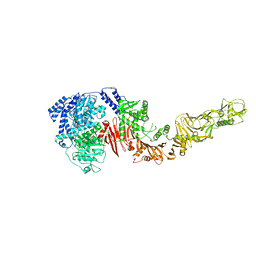 | |
6WA6
 
 | |
6WA9
 
 | |
5VQM
 
 | | Clostridium difficile TcdB-GTD bound to PA41 Fab | | Descriptor: | PA41 FAB HEAVY CHAIN, PA41 FAB LIGHT CHAIN, Toxin B | | Authors: | Kroh, H.K, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-05-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing antibody that blocks delivery of the enzymatic cargo of Clostridium difficile toxin TcdB into host cells.
J. Biol. Chem., 293, 2018
|
|
5UMI
 
 | | Clostridium difficile TcdA-CROPs bound to PA50 Fab | | Descriptor: | PA50 Fab Heavy chain, PA50 Fab Light chain, Toxin A | | Authors: | Kroh, H.K, Chandrasekaran, R, Rosenthal, K, Woods, R, Jin, X, Ohi, M.D, Nyborg, A.C, Rainey, G.J, Warrener, P, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-01-27 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Use of a neutralizing antibody helps identify structural features critical for binding of Clostridium difficile toxin TcdA to the host cell surface.
J. Biol. Chem., 292, 2017
|
|
3EB8
 
 | | VirA | | Descriptor: | Cysteine protease-like virA | | Authors: | Germane, K.L, Spiller, B.W. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules.
Biochemistry, 47, 2008
|
|
3SS1
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain | | Descriptor: | MANGANESE (II) ION, Toxin A | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
3SRZ
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain bound to UDP-glucose | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
