8UJW
 
 | |
1RTS
 
 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
3B5B
 
 | | Crystal structure of the thymidylate synthase k48q | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-(DIHYDROGEN PHOSPHATE), 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, FORMIC ACID, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of an invariant lysine residue in folate binding on Escherichia coli thymidylate synthase: Calorimetric and crystallographic analysis of the K48Q mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
8UZ7
 
 | | Crystal structure of a novel triose phosphate isomerase identified on the shrimp transcriptome | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Sotelo-Mundo, R.R, Gomez-Yanes, A.C, Lopez-Zavala, A.A, Lopez-Garcia, J.A, Ochoa-Leyva, A. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a novel triose phosphate isomerase from shrimp
To Be Published
|
|
1FFL
 
 | |
1FWM
 
 | | Crystal structure of the thymidylate synthase R166Q mutant | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Sotelo-Mundo, R.R, Changchien, L, Maley, F, Montfort, W.R. | | Deposit date: | 2000-09-23 | | Release date: | 2003-11-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thymidylate synthase mutant R166Q: Structural basis for the nearly complete loss of catalytic activity.
J.Biochem.Mol.Toxicol., 20, 2006
|
|
2VET
 
 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH DUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
2VF0
 
 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, SULFATE ION, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
2TSR
 
 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
5KEJ
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
7RE6
 
 | |
5AN1
 
 | |
7MIQ
 
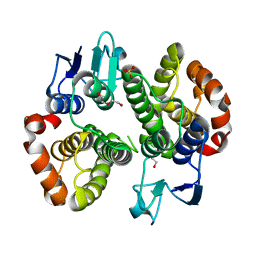 | | Crystal structure of a Glutathione S-transferase class Gtt2 of Vibrio parahaemolyticus (VpGSTT2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutathione S-transferase, SULFATE ION | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A.A, Garcia-Orozco, K.D, Sotelo-Mundo, R.R. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Novel Glutathione S -Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen.
Toxins, 13, 2021
|
|
5EYW
 
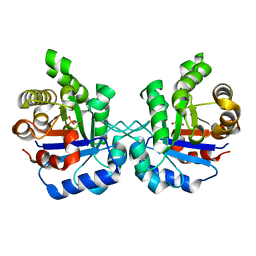 | | Crystal structure of Litopenaeus vannamei triosephosphate isomerase complexed with 2-Phosphoglycolic acid | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, Brieba, L.G, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2015-11-25 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights from a novel invertebrate triosephosphate isomerase from Litopenaeus vannamei.
Biochim.Biophys.Acta, 1864, 2016
|
|
5U92
 
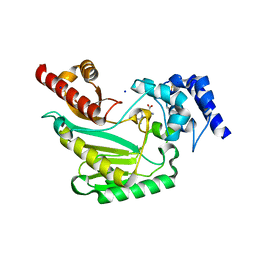 | | Crystal Structure of arginine kinase from the spider Polybetes pythagoricus in complex with arginine | | Descriptor: | ARGININE, SODIUM ION, arginine kinase | | Authors: | Lopez-zavala, A.A, Garcia, C.F, Paredes-Hernandez, J, Stojanoff, V, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
5U8E
 
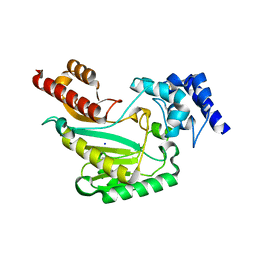 | | Crystal Structure of substrate-free arginine kinase from spider Polybetes pythagoricus | | Descriptor: | SODIUM ION, arginine kinase | | Authors: | Lopez-Zavala, A.A, Garcia, C.F, Hernadez-Paredes, J, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
4UOF
 
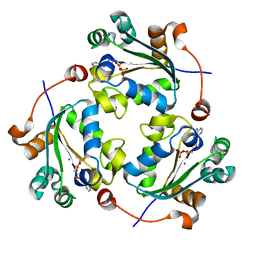 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dADP | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from the Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UOG
 
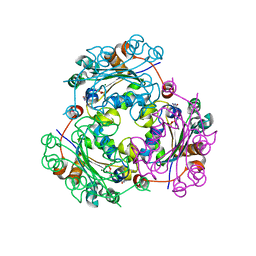 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UOH
 
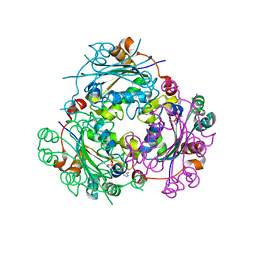 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Guevara-Hernandez, E, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
5G5E
 
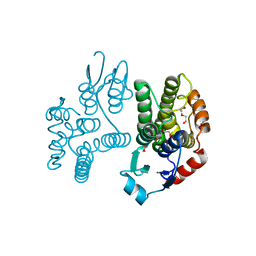 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU from mango Mangifera indica L. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
5G5F
 
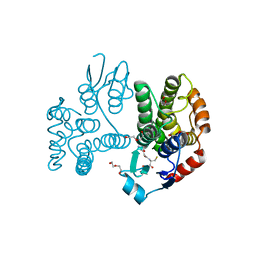 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with reduced glutathione. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
4CS5
 
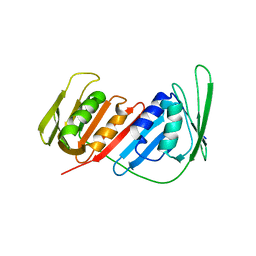 | | Crystal Structure of PCNA from Litopenaeus vannamei | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, De-La-Mora, E, Rudino-Pinera, E, Brieba, L.G, Sotelo-Mundo, R.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Shrimp Proliferating Cell Nuclear Antigen: Structural Complementarity with Wssv DNA Polymerase Pip-Box.
Plos One, 9, 2014
|
|
4BG4
 
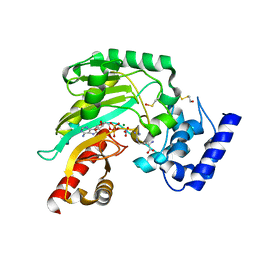 | | Crystal structure of Litopenaeus vannamei arginine kinase in a ternary analog complex with arginine, ADP-Mg and NO3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, ARGININE KINASE, ... | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, GBrieba, L, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-03-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
4BHL
 
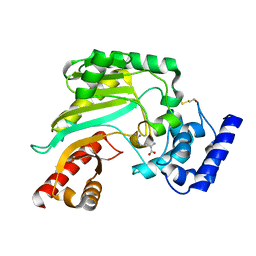 | | Crystal structure of Litopenaeus vannamei arginine kinase in binary complex with arginine | | Descriptor: | ARGININE, ARGININE KINASE, BETA-MERCAPTOETHANOL | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Hernandez-Flores, J.M, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, Brieba, L.G, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-04-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
4AM1
 
 | | Crystal structure of the marine crustacean decapod shrimp (Litopenaeus vannamei) arginine kinase in the absence of substrate or ligands. | | Descriptor: | ARGININE KINASE | | Authors: | Lopez-Zavala, A.A, Sotelo-Mundo, R.R, Garcia-Orozco, K.D, Isac-Martinez, F, Brieba, L.G, Rudino-Pinera, E. | | Deposit date: | 2012-03-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallization and X-Ray Diffraction Studies of Arginine Kinase from the White Pacific Shrimp Litopenaeus Vannamei.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
