3M3T
 
 | |
3M3S
 
 | |
5W0V
 
 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | 分子名称: | Histone H4 1-34, Kap123 | | 著者 | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | 登録日 | 2017-05-31 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.821 Å) | | 主引用文献 | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
4G88
 
 | | Crystal structure of OmpA peptidoglycan-binding domain from Acinetobacter baumannii | | 分子名称: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein Omp38, S,R MESO-TARTARIC ACID | | 著者 | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | 登録日 | 2012-07-22 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
4G4Z
 
 | |
4G4X
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | 分子名称: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | 著者 | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | 登録日 | 2012-07-16 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4GO6
 
 | | Crystal structure of HCF-1 self-association sequence 1 | | 分子名称: | HCF C-terminal chain 1, HCF N-terminal chain 1, SULFATE ION | | 著者 | Park, J, Lammers, F, Herr, W, Song, J. | | 登録日 | 2012-08-18 | | 公開日 | 2012-10-17 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | HCF-1 self-association via an interdigitated Fn3 structure facilitates transcriptional regulatory complex formation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5W40
 
 | |
5W3Y
 
 | |
2G31
 
 | | Human Nogo-A functional domain: nogo60 | | 分子名称: | Reticulon-4 | | 著者 | Li, M.F, Liu, J.X, Song, J.X. | | 登録日 | 2006-02-17 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration
Protein Sci., 15, 2006
|
|
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | 分子名称: | Beta-amyloid, Split Green flourescent protein | | 著者 | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | 登録日 | 2023-01-20 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
5IDH
 
 | |
5ICH
 
 | |
6YEJ
 
 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | 分子名称: | Huntingtin | | 著者 | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | 登録日 | 2020-03-24 | | 公開日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (18.200001 Å) | | 主引用文献 | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
7DEA
 
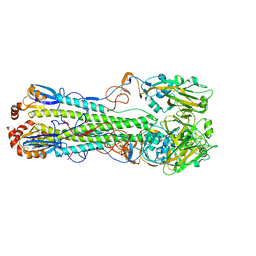 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | 登録日 | 2020-11-03 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
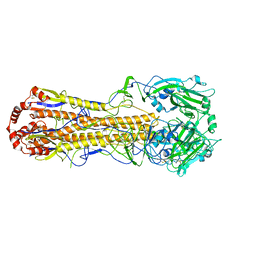 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | 登録日 | 2020-11-03 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
6F57
 
 | |
1HQY
 
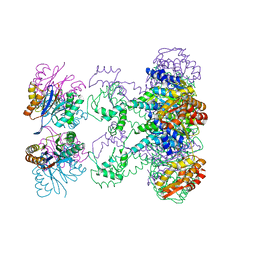 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | 著者 | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | 登録日 | 2000-12-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT1
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | 著者 | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | 登録日 | 2000-12-27 | | 公開日 | 2001-11-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | 著者 | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | 登録日 | 2000-12-27 | | 公開日 | 2001-11-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | 分子名称: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | 著者 | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | 登録日 | 2000-10-26 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (7 Å) | | 主引用文献 | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | 分子名称: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | 著者 | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | 登録日 | 2000-10-26 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
8EIH
 
 | |
8EII
 
 | |
8EIK
 
 | | Cryo-EM structure of human DNMT3B homo-hexamer | | 分子名称: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Lu, J.W, Song, J.K. | | 登録日 | 2022-09-15 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
