3KTJ
 
 | |
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
2AEH
 
 | | Focal adhesion kinase 1 | | 分子名称: | Focal adhesion kinase 1 | | 著者 | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | 登録日 | 2005-07-22 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
7XZ0
 
 | | TRIM E3 ubiquitin ligase | | 分子名称: | Tripartite motif-containing protein 72, ZINC ION | | 著者 | Park, S.H, Song, H.K. | | 登録日 | 2022-06-02 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2AL6
 
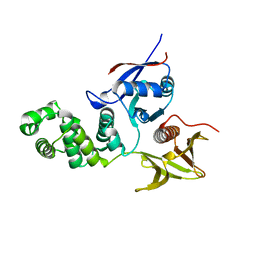 | | FERM domain of Focal Adhesion Kinase | | 分子名称: | Focal adhesion kinase 1 | | 著者 | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | 著者 | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | 登録日 | 2004-01-27 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
7Y6W
 
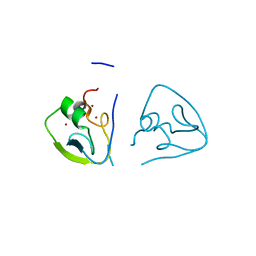 | | RRGSGG-AtPRT6 UBR box (I222) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | 分子名称: | Nucleoside Diphosphate Kinase A | | 著者 | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | 登録日 | 2001-09-10 | | 公開日 | 2002-04-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
5IZV
 
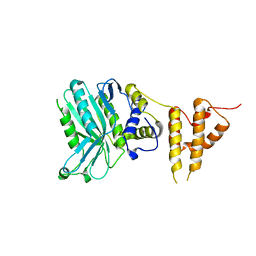 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-03-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
4XSJ
 
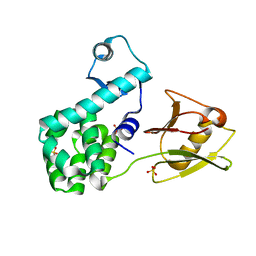 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | 分子名称: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | 著者 | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | 登録日 | 2015-01-22 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
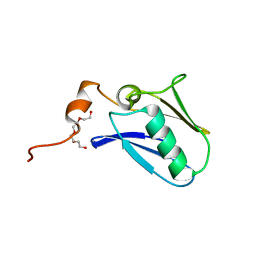 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | 分子名称: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | 著者 | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | 登録日 | 2015-01-23 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4TQ0
 
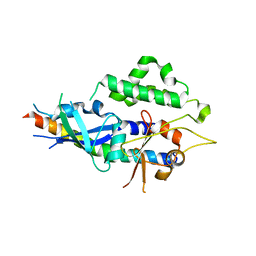 | |
4TQ1
 
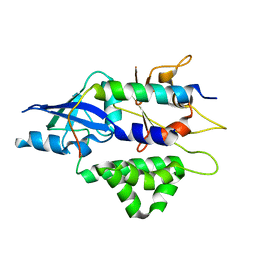 | | Crystal structure of human ATG5-TECAIR | | 分子名称: | Autophagy protein 5, Tectonin beta-propeller repeat-containing protein 1 | | 著者 | Kim, J.H, Hong, S.B, Song, H.K. | | 登録日 | 2014-06-10 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
2DS6
 
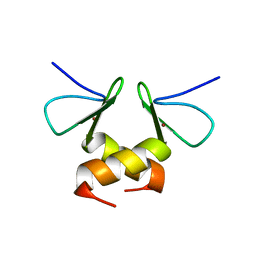 | | Structure of the ZBD in the tetragonal crystal form | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | 著者 | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | 登録日 | 2006-06-22 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS8
 
 | | Structure of the ZBD-XB complex | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | 著者 | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | 登録日 | 2006-06-22 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS7
 
 | | Structure of the ZBD in the hexagonal crystal form | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | 著者 | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | 登録日 | 2006-06-22 | | 公開日 | 2007-02-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1OIL
 
 | | STRUCTURE OF LIPASE | | 分子名称: | CALCIUM ION, LIPASE | | 著者 | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 1996-12-06 | | 公開日 | 1997-05-15 | | 最終更新日 | 2018-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1M27
 
 | | Crystal structure of SAP/FynSH3/SLAM ternary complex | | 分子名称: | CITRATE ANION, Proto-oncogene tyrosine-protein kinase FYN, SH2 domain protein 1A, ... | | 著者 | Chan, B, Griesbach, J, Song, H.K, Poy, F, Terhorst, C, Eck, M.J. | | 登録日 | 2002-06-21 | | 公開日 | 2003-05-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | SAP couples Fyn to SLAM immune receptors.
NAT.CELL BIOL., 5, 2003
|
|
1NU2
 
 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | 著者 | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | 登録日 | 2003-01-30 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NTV
 
 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | 分子名称: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | 著者 | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | 登録日 | 2003-01-30 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NHL
 
 | |
5JST
 
 | | MBP fused MDV1 coiled coil | | 分子名称: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein,Mitochondrial division protein 1, ... | | 著者 | Kim, B.-W, Song, H.K. | | 登録日 | 2016-05-09 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | ACCORD: an assessment tool to determine the orientation of homodimeric coiled-coils.
Sci Rep, 7, 2017
|
|
2H8G
 
 | | 5'-Methylthioadenosine Nucleosidase from Arabidopsis thaliana | | 分子名称: | 5'-Methylthioadenosine Nucleosidase, ADENINE | | 著者 | Park, E.Y, Oh, S.I, Nam, M.J, Shin, J.S, Kim, K.N, Song, H.K. | | 登録日 | 2006-06-07 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution
Proteins, 65, 2006
|
|
5K5U
 
 | |
