7GII
 
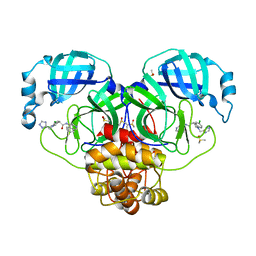 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-THA-8416115c-13 (Mpro-P0091) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-[(4H-1,2,4-triazol-3-yl)methyl]-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLJ
 
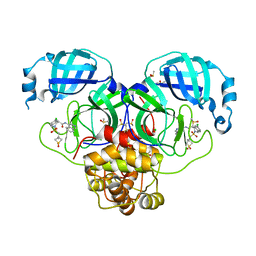 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-4fff0a85-3 (Mpro-P1983) | | 分子名称: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3-methyl-1,1-dioxo-1lambda~6~-thietan-3-yl)methanesulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.814 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GIY
 
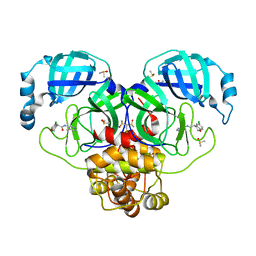 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-CON-c4e3408a-1 (Mpro-P0145) | | 分子名称: | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLZ
 
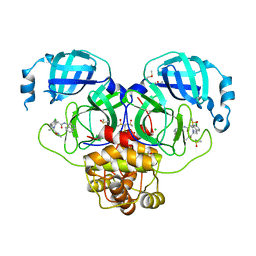 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-5cd9ea36-14 (Mpro-P2067) | | 分子名称: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(2S)-1-(methylamino)-1-oxopropan-2-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.678 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e69ad64a-2 (Mpro-P0213) | | 分子名称: | (3S)-5-chloro-N-(isoquinolin-4-yl)-N-propanoyl-2,3-dihydro-1-benzofuran-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GMG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e119ab4f-2 (Mpro-P2182) | | 分子名称: | (4S)-6-chloro-N-(6-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e9e99895-6 (Mpro-P0655) | | 分子名称: | (2R)-2-[2-(3-cyclopropyl-2-oxoimidazolidin-1-yl)acetamido]-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3KWO
 
 | | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni | | 分子名称: | 1,4-BUTANEDIOL, ACETIC ACID, GLYCEROL, ... | | 著者 | Kim, Y, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-12-01 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.985 Å) | | 主引用文献 | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni
To be Published
|
|
3LXY
 
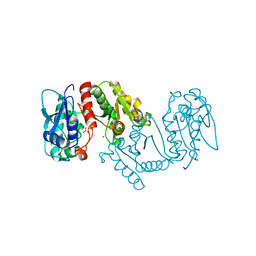 | | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92 | | 分子名称: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Nocek, B, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-02-25 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92
To be Published
|
|
3LUS
 
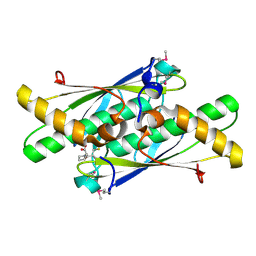 | | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961 | | 分子名称: | L-CAPTOPRIL, Organic hydroperoxide resistance protein | | 著者 | Nocek, B, Maltseva, N, Makowska-Grzyska, M, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-02-18 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961
TO BE PUBLISHED
|
|
3JR2
 
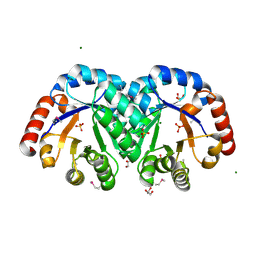 | | X-ray crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Hexulose-6-phosphate synthase SgbH, ... | | 著者 | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-09-08 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate
decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3M34
 
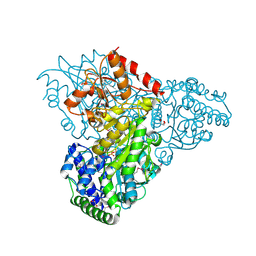 | | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-03-08 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion
To be Published
|
|
3M7I
 
 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion | | 分子名称: | 1,2-ETHANEDIOL, 5-O-phosphono-beta-D-ribofuranose, MAGNESIUM ION, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-03-16 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion
TO BE PUBLISHED
|
|
3M6L
 
 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-03-15 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion
To be Published
|
|
7X5L
 
 | | Tir-dsDNA complex, the initial binding state | | 分子名称: | DNA (5'-D(*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*AP*TP*TP*AP*A)-3'), Flax rust resistance protein | | 著者 | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | 登録日 | 2022-03-04 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
7X5M
 
 | | Tir-dsDNA complex, the initial binding state | | 分子名称: | 2',3'- cyclic AMP, DNA (5'-D(P*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*AP*TP*TP*TP*A)-3'), ... | | 著者 | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | 登録日 | 2022-03-05 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
7X5K
 
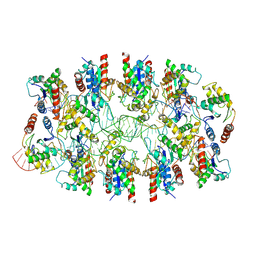 | | Tir-dsDNA complex, the initial binding state | | 分子名称: | DNA (43-MER), Flax rust resistance protein | | 著者 | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | 登録日 | 2022-03-04 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
5ROB
 
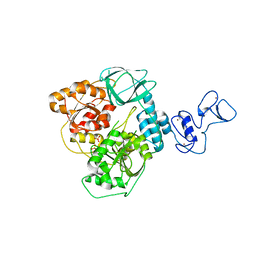 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 helicase | | 分子名称: | Helicase, PHOSPHATE ION, ZINC ION | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-22 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5S73
 
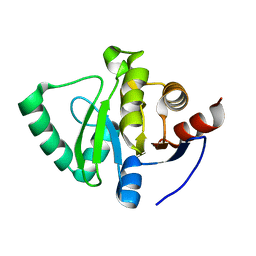 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
5S74
 
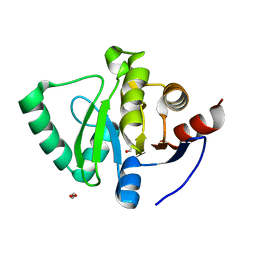 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-23 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
5WRG
 
 | | SARS-CoV spike glycoprotein | | 分子名称: | Spike glycoprotein | | 著者 | Gui, M, Song, W, Xiang, Y, Wang, X. | | 登録日 | 2016-12-01 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
5XLR
 
 | |
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | 著者 | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-07-15 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
3P7M
 
 | | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | Malate dehydrogenase, PHOSPHATE ION | | 著者 | Osinski, T, Cymborowski, M, Zimmerman, M.D, Gordon, E, Grimshaw, S, Skarina, T, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-10-12 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | 分子名称: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | 著者 | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | 登録日 | 2020-03-06 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
