6Z07
 
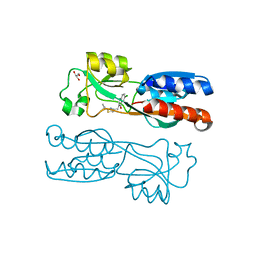 | | PqsR (MvfR) in complex with antagonist 12 | | Descriptor: | 3-[(2-~{tert}-butyl-1,3-thiazol-4-yl)methyl]-6-chloranyl-quinazolin-4-one, GLYCEROL, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
8A8E
 
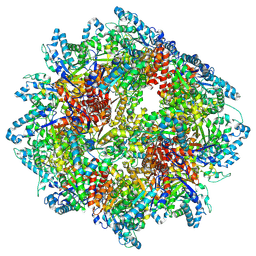 | | PPSA C terminal octahedral structure | | Descriptor: | Phosphoenolpyruvate synthase | | Authors: | Song, W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase
To Be Published
|
|
5XZJ
 
 | |
6Z5K
 
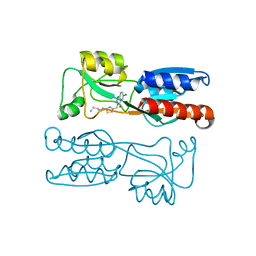 | | PqsR (MvfR) in complex with antagonist 18 | | Descriptor: | 6-chloranyl-3-[(2-pentyl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
7C7Z
 
 | |
5XZI
 
 | |
8H2C
 
 | | Crystal structure of the pseudaminic acid synthase PseI from Campylobacter jejuni | | Descriptor: | MANGANESE (II) ION, Pseudaminic acid synthase | | Authors: | Song, W.S, Park, M.A, Ki, D.U, Yoon, S.I. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7DCL
 
 | |
5HZ3
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGFR5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-PRO-LYS-PRO-SER-THR-ARG-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HZ1
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-ARG-ALA-LYS-HIS-HIS-PRO-HYP-LYS-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Plant Receptor
To Be Published
|
|
4XPK
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH | | Descriptor: | N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
4XPL
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
6MPV
 
 | |
5CC1
 
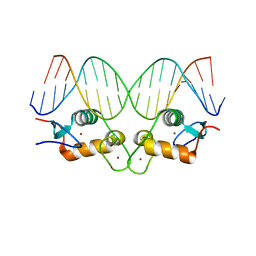 | | S425G Glucocorticoid receptor DNA binding domain - (+)GRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Weikum, E.A, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBX
 
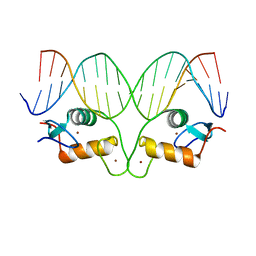 | | AncGR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBZ
 
 | | AncMR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncMR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1CRN
 
 | |
5CC0
 
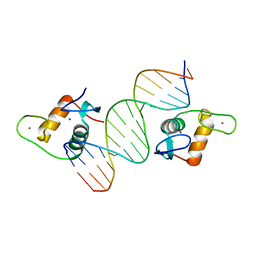 | | AncSR2 - TSLP nGRE complex | | Descriptor: | AncSR2 DNA Binding Domain, DNA (5'-D(*AP*GP*CP*TP*CP*TP*CP*CP*CP*GP*GP*AP*GP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*CP*TP*CP*CP*GP*GP*GP*AP*GP*AP*GP*CP*T)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBY
 
 | | AncGR2 DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR2 DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6AI5
 
 | | Disulfide-free, Zn-directed tetramer of the engineered cyt cb562 variant, C96T/A104AB3 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Song, W.J, Tezcan, F.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Disulfide-free, Zn-directed tetramer of the engineered cyt cb562 variant, C96T/A104AB3
To Be Published
|
|
8YLF
 
 | |
8YLM
 
 | |
8YLJ
 
 | |
8YLH
 
 | |
