7SIM
 
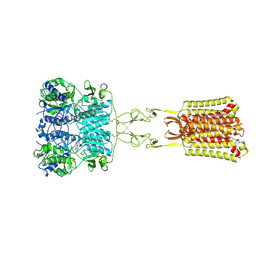 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3V05
 
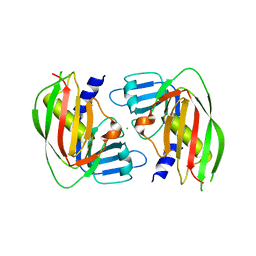 | | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Superantigen-like Protein | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Filippova, E.V, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus.
TO BE PUBLISHED
|
|
4FIT
 
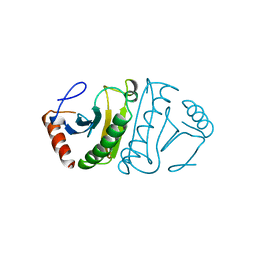 | | FHIT-APO | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
3UWQ
 
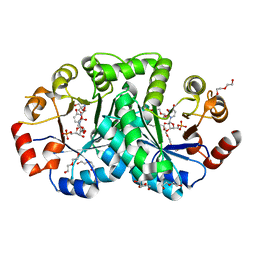 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
4GIB
 
 | | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile | | Descriptor: | Beta-phosphoglucomutase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile.
TO BE PUBLISHED
|
|
2C2A
 
 | |
6CTY
 
 | | Crystal structure of dihydroorotase pyrC from Yersinia pestis in complex with zinc and malate at 2.4 A resolution | | Descriptor: | D-MALATE, Dihydroorotase, ZINC ION | | Authors: | Lipowska, J, Shabalin, I.G, Winsor, J, Woinska, M, Cooper, D.R, Kwon, K, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
7O2U
 
 | | PqsR (MvfR) in complex with antagonist 40 | | Descriptor: | 2-[4-[(2S)-3-(6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanyl-propoxy]phenoxy]ethanenitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 40
To Be Published
|
|
3UY4
 
 | | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5
To be Published
|
|
1IEB
 
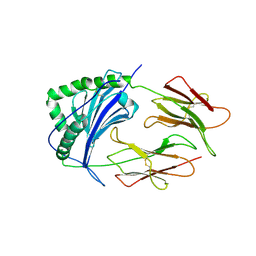 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK, SULFATE ION | | Authors: | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | Deposit date: | 1996-04-05 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
7O2T
 
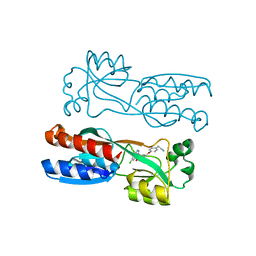 | | PqsR (MvfR) in complex with antagonist 61 | | Descriptor: | 2-[4-[(2S)-3-(6-chloro-4-oxoquinazolin-3-yl)-2-hydroxypropoxy]phenyl]acetonitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 61
To Be Published
|
|
3USB
 
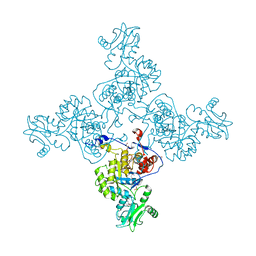 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
5KP2
 
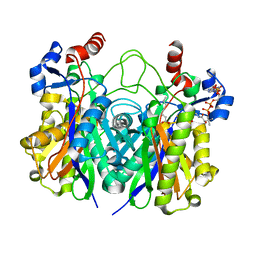 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
5DN8
 
 | | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP. | | Descriptor: | GTPase Der, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Minasov, G, Shuvalova, L, Han, A, Kim, H.-Y, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.
To Be Published
|
|
3UXV
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANINE, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ
To be Published, 2012
|
|
3UPB
 
 | | 1.5 Angstrom Resolution Crystal Structure of Transaldolase from Francisella tularensis in Covalent Complex with Arabinose-5-Phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Arabinose 5-phosphate covalently inhibits transaldolase.
J.Struct.Funct.Genom., 15, 2014
|
|
4GFS
 
 | | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site | | Descriptor: | 3-dehydroquinate dehydratase, NICKEL (II) ION, SUCCINIC ACID | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site
TO BE PUBLISHED
|
|
3LAY
 
 | | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Zinc resistance-associated protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
5KJP
 
 | |
3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, GLYCEROL, Transketolase | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-29 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
6CN1
 
 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
3LER
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-15 | | Release date: | 2010-01-26 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4WD7
 
 | |
3LB0
 
 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, CITRIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site.
TO BE PUBLISHED
|
|
