6VFB
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Reaction State Ternary Complex, 50 mM Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Reaction State Ternary Complex, 50 mM Mn2+ (960 min)
To be published
|
|
6VFC
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min)
To be published
|
|
6N2S
 
 | |
6N2T
 
 | |
6NKV
 
 | |
6NKX
 
 | |
6NKR
 
 | | Ternary complex crystal structure of K289M variant of DNA polymerase Beta with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Revealing an Internal Stabilization Deficiency in the DNA Polymerase beta K289M Cancer Variant through the Combined Use of Chemical Biology and X-ray Crystallography.
Biochemistry, 59, 2020
|
|
6NKU
 
 | |
6NKS
 
 | |
6NKW
 
 | |
6NL0
 
 | |
7UYU
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYR
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 12 | | Descriptor: | 2-(4-{[2-(2,6-difluorophenyl)-5-oxo-5H-pyrrolo[3,4-d]pyrimidin-4-yl]amino}phenyl)-N-ethylacetamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYW
 
 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
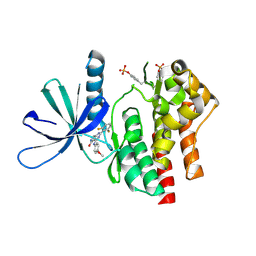 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
6NKZ
 
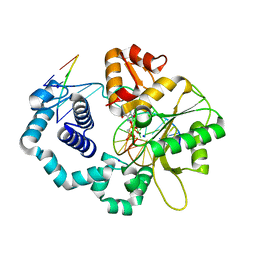 | |
6NKT
 
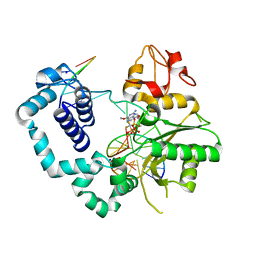 | |
6NKY
 
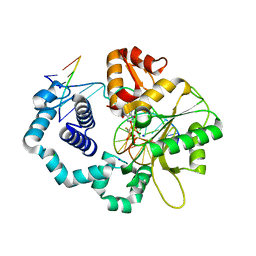 | |
6W2M
 
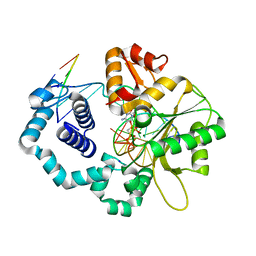 | |
2Y7Z
 
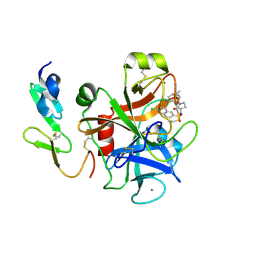 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-[(1S)-1-DIMETHYLAMINO-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y82
 
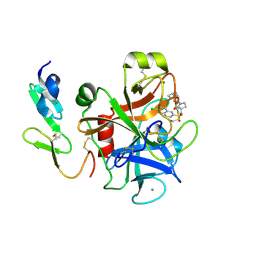 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2S)-2-PYRROLIDINYL]PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y81
 
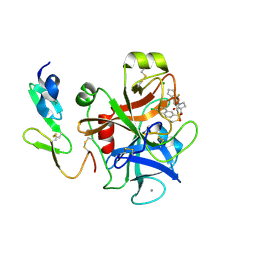 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2R)-2--PYRROLIDINYL] PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y80
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
