7OEA
 
 | | Lassa virus L protein bound to 3' promoter RNA (well-resolved polymerase core) [3END-CORE] | | Descriptor: | 3' vRNA, MAGNESIUM ION, RNA-directed RNA polymerase L, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OE3
 
 | | Apo-structure of Lassa virus L protein (well-resolved endonuclease) [APO-ENDO] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OEB
 
 | | Lassa virus L protein bound to 3' promoter RNA (well-resolved endonuclease) [3END-ENDO] | | Descriptor: | 3' vRNA, MAGNESIUM ION, RNA-directed RNA polymerase L, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OCH
 
 | | Apo-structure of Lassa virus L protein (well-resolved polymerase core) [APO-CORE] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
1DDK
 
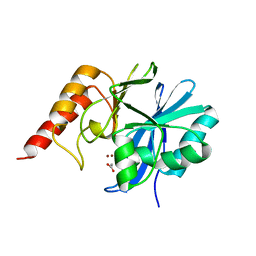 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DD6
 
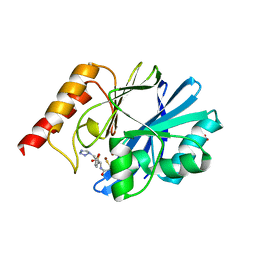 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1MOF
 
 | | COAT PROTEIN | | Descriptor: | CHLORIDE ION, MOLONEY MURINE LEUKEMIA VIRUS P15 | | Authors: | Fass, D, Harrison, S.C, Kim, P.S. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retrovirus envelope domain at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
8FNT
 
 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8V7S
 
 | | IpaD (122-321) Apo Structure | | Descriptor: | Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J, Morales, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
2BNA
 
 | |
1INQ
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | Descriptor: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
8AX6
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029882 | | Descriptor: | (1~{S},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8AX7
 
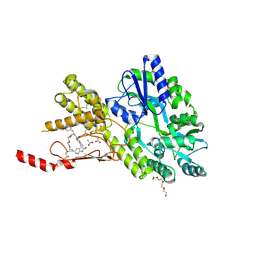 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0031448 | | Descriptor: | (1~{S},10~{R},20~{E})-10-[(1,7-dimethylindazol-5-yl)methyl]-12-methyl-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, ACETATE ION, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
1KTA
 
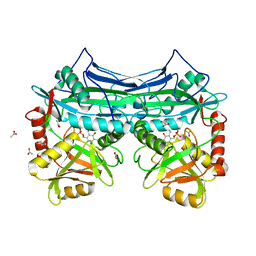 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE : THREE DIMENSIONAL STRUCTURE OF THE ENZYME IN ITS PYRIDOXAMINE PHOSPHATE FORM. | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETIC ACID, ... | | Authors: | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | Deposit date: | 2002-01-15 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
1KT8
 
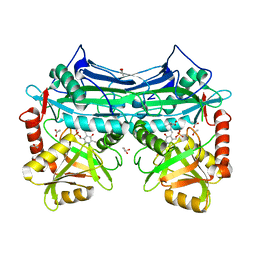 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME IN ITS KETIMINE FORM WITH THE SUBSTRATE L-ISOLEUCINE | | Descriptor: | ACETIC ACID, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, MITOCHONDRIAL, ... | | Authors: | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | Deposit date: | 2002-01-15 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
8THU
 
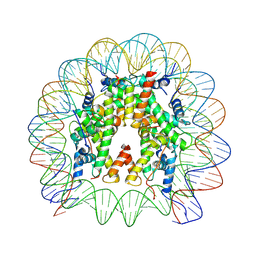 | |
8T9H
 
 | |
3DZC
 
 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
2WO9
 
 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-(4'-ACETYLBIPHENYL-4-YL)-3-HYDROXYPENTANOIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5EQ2
 
 | | Crystal Structure of the SrpA Adhesin from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
1S9N
 
 | | Solution structure of the nitrous acid (G)-(G) cross-linked DNA dodecamer duplex GCATCC(G)GATGC | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*CP*GP*GP*AP*TP*GP*C)-3' | | Authors: | Edfeldt, N.B.F, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link
Nucleic Acids Res., 32, 2004
|
|
8V5C
 
 | | IpaD (122-321) Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2023-11-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
7LV9
 
 | | Marseillevirus heterotrimeric (hexameric) nucleosome | | Descriptor: | DNA (96-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LV8
 
 | | Structure of the Marseillevirus nucleosome | | Descriptor: | DNA (123-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
