6PL6
 
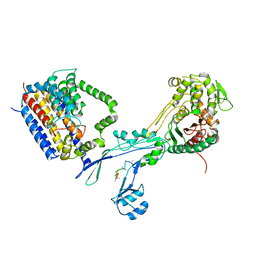 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | 著者 | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6POS
 
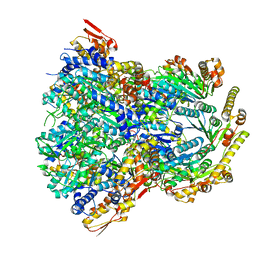 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO1
 
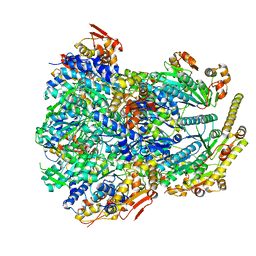 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP8
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
7OJ8
 
 | | ABCG2 E1S turnover-2 state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL, ... | | 著者 | Ni, D, Yu, Q, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | 登録日 | 2021-05-14 | | 公開日 | 2021-07-21 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
4RT3
 
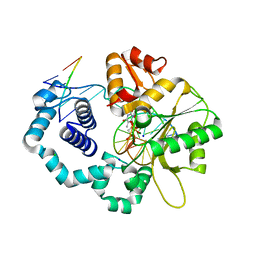 | | Ternary complex crystal structure of DNA polymerase Beta with (alpha, beta)-NH-(beta,gamma)-CH2-dTTP | | 分子名称: | 2'-deoxy-5'-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]amino}phosphoryl]-3,4-dihydrothymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | 著者 | Batra, V.K, Wilson, S.H. | | 登録日 | 2014-11-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Two Scaffolds from Two Flips: (alpha, beta)/(beta, gamma) CH2/NH "Met-Im" Analogues of dTTP.
Org.Lett., 17, 2015
|
|
5CFP
 
 | | Crystal structure of anemone STING (Nematostella vectensis) 'humanized' F276K in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p]' | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.066 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5CFO
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'rotated' open conformation | | 分子名称: | Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5CFR
 
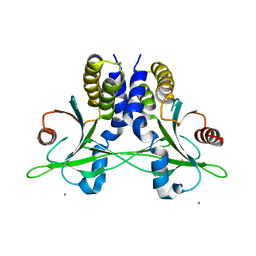 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'unrotated' closed conformation | | 分子名称: | CALCIUM ION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
6YCL
 
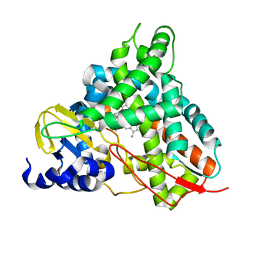 | | Crystal structure of GcoA T296G bound to p-vanillin | | 分子名称: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | 著者 | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | 分子名称: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | 著者 | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCP
 
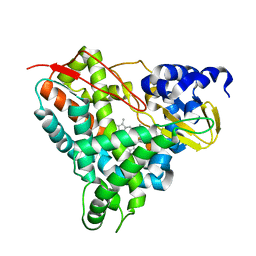 | | Crystal structure of GcoA F169V bound to o-vanillin | | 分子名称: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | 著者 | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
4TT9
 
 | |
6YCN
 
 | | Crystal structure of GcoA F169A bound to o-vanillin | | 分子名称: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | 著者 | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCI
 
 | | Crystal structure of GcoA T296G bound to guaiacol | | 分子名称: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | 著者 | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
5CFM
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' cGAMP, c[G(3', 5')pA(3', 5')p] | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CITRATE ANION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
6T26
 
 | | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine | | 分子名称: | Alkaline phosphatase, CHLORIDE ION, CYCLOHEXYLAMMONIUM ION, ... | | 著者 | Asgeirsson, B, Hjorleifsson, J.G, Markusson, S, Helland, R. | | 登録日 | 2019-10-07 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.265 Å) | | 主引用文献 | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Biochem Biophys Rep, 24, 2020
|
|
4RM4
 
 | | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhouw, W.H, Zhang, A.L, Zhang, T, Hall, E.A, Hutchinson, S, Cryle, M.J, Wong, L.-L, Bell, S.G. | | 登録日 | 2014-10-18 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.771 Å) | | 主引用文献 | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis.
Mol Biosyst, 11, 2015
|
|
6YSL
 
 | |
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | 分子名称: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | 著者 | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-03-05 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
6G2Q
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (S-isomer), beta, gamma dTTP analogue | | 分子名称: | CHLORIDE ION, DNA polymerase beta, Downstream Primer Strand, ... | | 著者 | Batra, V.K, Wilson, S.H. | | 登録日 | 2018-03-23 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.148 Å) | | 主引用文献 | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
5DFF
 
 | | Human APE1 product complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Freudenthal, B.D, Wilson, S.H. | | 登録日 | 2015-08-26 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFH
 
 | | Human APE1 mismatch product complex | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | 著者 | Freudenthal, B.D, Wilson, S.H. | | 登録日 | 2015-08-26 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
