3LS1
 
 | | Crystal Structure of Cyanobacterial PsbQ from Synechocystis sp. PCC 6803 complexed with Zn2+ | | Descriptor: | Sll1638 protein, ZINC ION | | Authors: | Jackson, S.A, Fagerlund, R.D, Wilbanks, S.M, Eaton-Rye, J.J. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: Implications for Binding and Function in Cyanobacterial Photosystem II
Biochemistry, 49, 2010
|
|
3SCW
 
 | | Crystal Structure of Rice BGlu1 E386G/Y341A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3LS0
 
 | |
7R20
 
 | | Anti-Arc nanobody E5 | | Descriptor: | Anti-Arc nanobody E5, GLYCEROL, SULFATE ION | | Authors: | Markusson, S, Kursula, P. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-affinity anti-Arc nanobodies provide tools for structural and functional studies.
Plos One, 17, 2022
|
|
3SCQ
 
 | | Crystal Structure of Rice BGlu1 E386A Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCP
 
 | | Crystal Structure of Rice BGlu1 E386A Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCS
 
 | | Crystal Structure of Rice BGlu1 E386S Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
7UWL
 
 | | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
3G0Y
 
 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | Descriptor: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6YI5
 
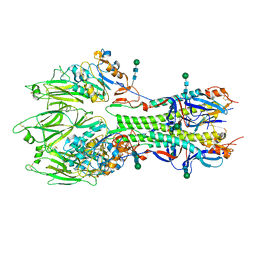 | | In-situ structure of the trimeric HEF from influenza C by flexible fitting into a cryo-ET map. | | Descriptor: | Hemagglutinin-esterase-fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Halldorsson, S, Rosenthal, P.B. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | In situ structure and organization of the influenza C virus surface glycoprotein.
Nat Commun, 12, 2021
|
|
3D0L
 
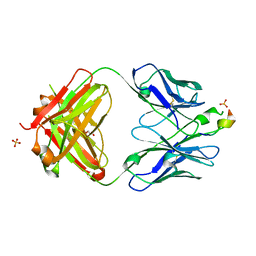 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 | | Descriptor: | 2F5 heavy chain, 2F5 light chain, GLYCEROL, ... | | Authors: | Bryson, S, Julien, J.P, Pai, E.F. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3G11
 
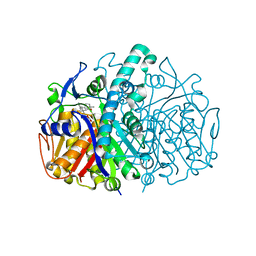 | | Structure of E. coli FabF(C163Q) in complex with dihydrophenyl platensimycin | | Descriptor: | 3-({3-[(1S,4S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxo-4-phenyldecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6BU7
 
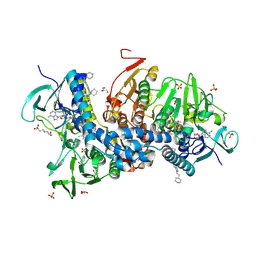 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD130 1-[2-(Piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-08 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
7AKV
 
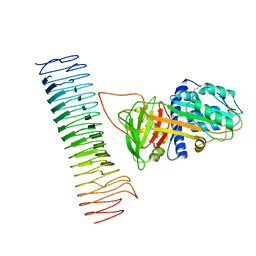 | | The cryo-EM structure of the Vag8-C1 inhibitor complex | | Descriptor: | Plasma protease C1 inhibitor, Vag8 | | Authors: | Johnson, S, Lea, S.M, Deme, J.C, Furlong, E, Dhillon, A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Mbio, 12, 2021
|
|
7UPM
 
 | |
7BHQ
 
 | |
6BTL
 
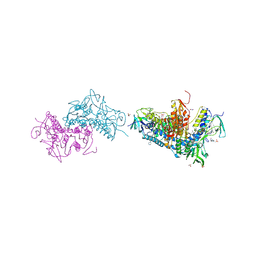 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD117 1-[2-(Piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
3N2Z
 
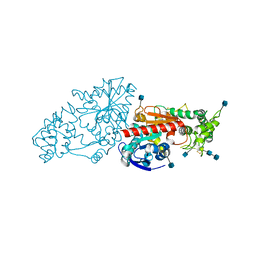 | | The Structure of Human Prolylcarboxypeptidase at 2.80 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal Pro-X carboxypeptidase, ... | | Authors: | Soisson, S.M, Patel, S.B, Lumb, K.J, Sharma, S. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural definition and substrate specificity of the S28 protease family: the crystal structure of human prolylcarboxypeptidase.
Bmc Struct.Biol., 10, 2010
|
|
6HIJ
 
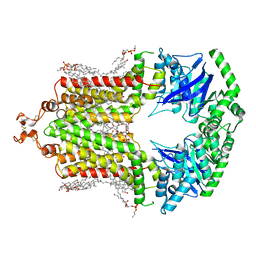 | | Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATP-binding cassette sub-family G member 2, CHOLESTEROL, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-08-30 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat.Struct.Mol.Biol., 25, 2018
|
|
2TBV
 
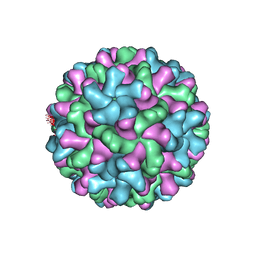 | |
6YCH
 
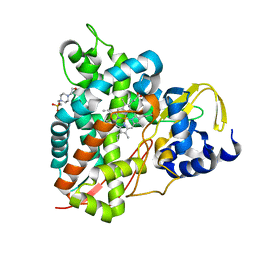 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCM
 
 | | Crystal structure of GcoA T296S bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCJ
 
 | | Crystal structure of GcoA T296S bound to guaiacol | | Descriptor: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7BZM
 
 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
7BK0
 
 | |
