6URN
 
 | | Barrier-to-autointegration factor t-butanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1PIH
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
9CYO
 
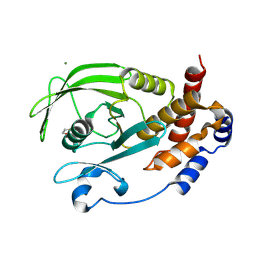 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYQ
 
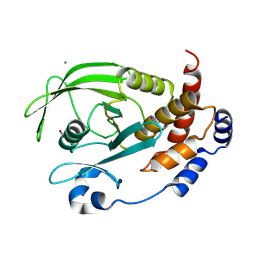 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYR
 
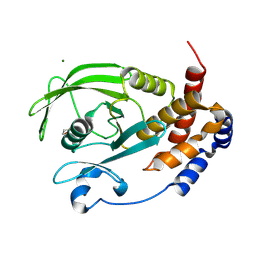 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYP
 
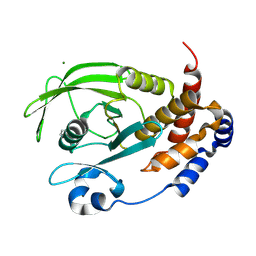 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
1NZ2
 
 | | K45E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, M, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6MYN
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to inhibitor R7 | | Descriptor: | (5s,7s)-9-fluoro-10-[(3R)-3-hydroxy-3-(5-methyl-1,2-oxazol-3-yl)but-1-yn-1-yl]-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Harris, S.F, Smith, M, Barker, J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
3EV3
 
 | | Crystal Structure of Ribonuclease A in 70% t-Butanol | | Descriptor: | Ribonuclease pancreatic, TERTIARY-BUTYL ALCOHOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV0
 
 | | Crystal Structure of Ribonuclease A in 70% Dimethyl Sulfoxide | | Descriptor: | DIMETHYL SULFOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUY
 
 | | Crystal Structure of Ribonuclease A in 50% Dioxane | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV6
 
 | | Crystal Structure of Ribonuclease A in 50% R,S,R-Bisfuranol | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUX
 
 | | Crystal Structure of Crosslinked Ribonuclease A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV4
 
 | | Crystal Structure of Ribonuclease A in 50% Trifluoroethanol | | Descriptor: | Ribonuclease pancreatic, TRIFLUOROETHANOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUZ
 
 | | Crystal Structure of Ribonuclease A in 50% Dimethylformamide | | Descriptor: | DIMETHYLFORMAMIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV1
 
 | | Crystal Structure of Ribonuclease A in 70% Hexanediol | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV2
 
 | | Crystal Structure of Ribonuclease A in 70% Isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV5
 
 | | Crystal Structure of Ribonuclease A in 1M Trimethylamine N-Oxide | | Descriptor: | Ribonuclease pancreatic, trimethylamine oxide | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
6O2E
 
 | |
6O2F
 
 | |
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
5HQ3
 
 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
