5LOF
 
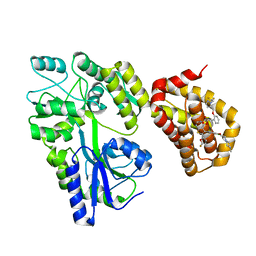 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
4XVZ
 
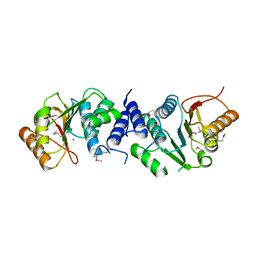 | | MycF mycinamicin III 3'-O-methyltransferase in complex with Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
7UCH
 
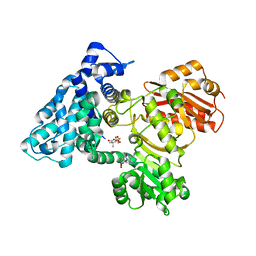 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Di-methyl-malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Skiba, M.A, Lao, Y, Smith, J.L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Basis for Control of Methylation Extent in Polyketide Synthase Metal-Dependent C -Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
5K6K
 
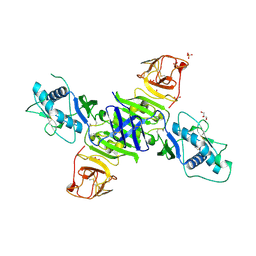 | | Zika virus non-structural protein 1 (NS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Akey, D.L, Brown, W.C, Smith, J.L. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Extended surface for membrane association in Zika virus NS1 structure.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6ECX
 
 | | StiE O-MT residues 942-1257 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.B, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6MBG
 
 | |
6MBF
 
 | | GphF Dehydratase 1 | | Descriptor: | GphF Dehydratase 1, MAGNESIUM ION | | Authors: | Dodge, G.J, Smith, J.L. | | Deposit date: | 2018-08-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Molecular Basis for Olefin Rearrangement in the Gephyronic Acid Polyketide Synthase.
ACS Chem. Biol., 13, 2018
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
1OX6
 
 | |
1OX4
 
 | |
6MBH
 
 | |
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL8
 
 | | Crystal structure HpiC1 Y101F/F138S | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6NET
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVH
 
 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVG
 
 | | Crystal structure of ligand free PhqK | | Descriptor: | FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVI
 
 | | Crystal structure of PhqK in complex with paraherquamide L | | Descriptor: | (8aS,13S,13aR,14aS)-4,4,13,15,15-pentamethyl-12,13,14,14a,15,16-hexahydro-4H,8H,9H,11H-8a,13a-(epiminomethano)[1,4]dioxepino[2,3-a]indolizino[6,7-h]carbazol-17-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6NES
 
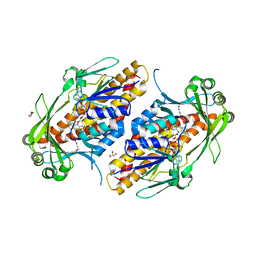 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6ZHD
 
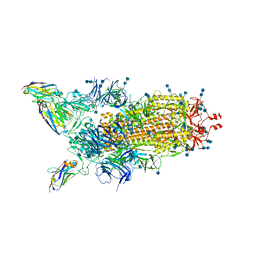 | | H11-H4 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H4, ... | | Authors: | Clare, D.K, Naismith, J.H, Weckener, M, Vogirala, V.K. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | H11-H4 bound to Spike
To Be Published
|
|
5LDT
 
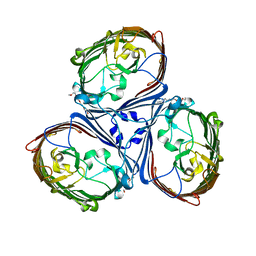 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | Authors: | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
6PVJ
 
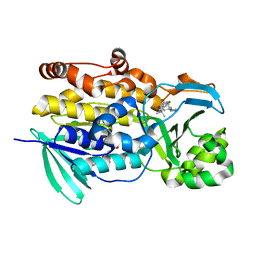 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6ECV
 
 | | StiD O-MT residues 976-1266 | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECT
 
 | | StiE O-MT residues 961-1257 | | Descriptor: | S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
