6I3L
 
 | | Bilirubin oxidase from Myrothecium verrucaria, mutant W396F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Koval, T, Svecova, L, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
1IIQ
 
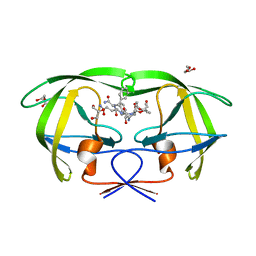 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE COMPLEXED WITH A HYDROXYETHYLAMINE PEPTIDOMIMETIC INHIBITOR | | Descriptor: | GLYCEROL, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Dohnalek, J, Hasek, J, Duskova, J, Petrokova, H, Hradilek, M, Soucek, M, Konvalinka, J, Brynda, J, Sedlacek, J, Fabry, M. | | Deposit date: | 2001-04-24 | | Release date: | 2002-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Hydroxyethylamine isostere of an HIV-1 protease inhibitor prefers its amine to the hydroxy group in binding to catalytic aspartates. A synchrotron study of HIV-1 protease in complex with a peptidomimetic inhibitor.
J.Med.Chem., 45, 2002
|
|
1SP5
 
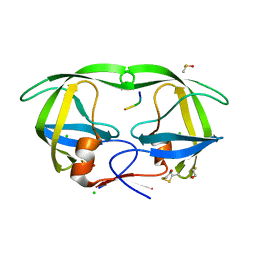 | | Crystal structure of HIV-1 protease complexed with a product of autoproteolysis | | Descriptor: | 5-mer peptide from Protease, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Vondrackova, E, Hasek, J, Jaskolski, M, Rezacova, P, Dohnalek, J, Skalova, T, Petrokova, H, Duskova, J, Brynda, J, Sedlacek, J. | | Deposit date: | 2004-03-16 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Product of enzymatic self-cleavage bound in the active site of HIV protease
To be Published
|
|
1FQX
 
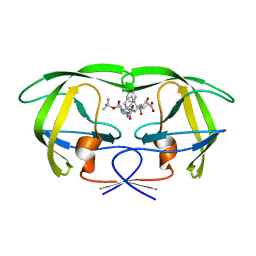 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HIV-1 PROTEASE WITH A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Dohnalek, J, Hasek, J, Duskova, J, Petrokova, H, Hradilek, M, Soucek, M, Konvalinka, J, Brynda, J, Sedlacek, J, Fabry, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A distinct binding mode of a hydroxyethylamine isostere inhibitor of HIV-1 protease.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ZJ7
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
1ZLF
 
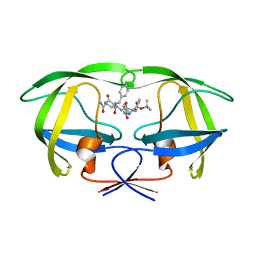 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
4QKH
 
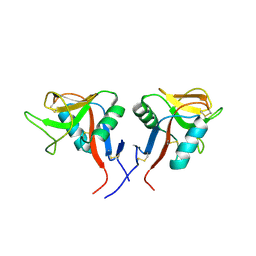 | | Dimeric form of human LLT1, a ligand for NKR-P1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D | | Authors: | Skalova, T, Blaha, J, Harlos, K, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QKI
 
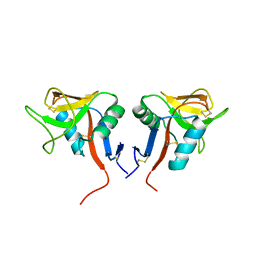 | | Dimeric form of human LLT1, a ligand for NKR-P1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D | | Authors: | Skalova, T, Blaha, J, Harlos, K, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QKG
 
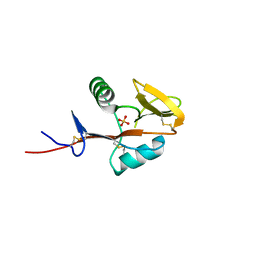 | | Monomeric form of human LLT1, a ligand for NKR-P1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, SULFATE ION | | Authors: | Skalova, T, Blaha, J, Harlos, K, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QKJ
 
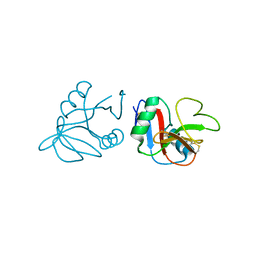 | | Glycosylated form of human LLT1, a ligand for NKR-P1, in this structure forming hexamers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D | | Authors: | Skalova, T, Blaha, J, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RGS
 
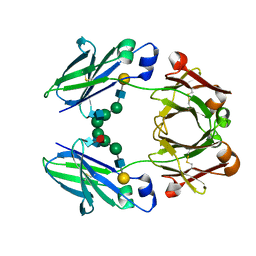 | | FC-fragment of monoclonal antibody IGG2B from Mus musculus | | Descriptor: | Ig gamma-2B heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J, Hasek, J. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution
Immunology, 126, 2008
|
|
3KW8
 
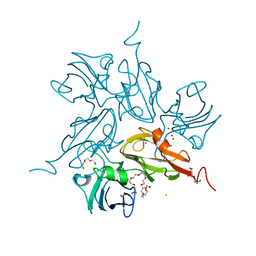 | | Two-domain laccase from Streptomyces coelicolor at 2.3 A resolution | | Descriptor: | COPPER (II) ION, FE (III) ION, Putative copper oxidase, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Duskova, J, Stepankova, A, Hasek, J, Ostergaard, L.H, Ostergaard, P.R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 A
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3SNG
 
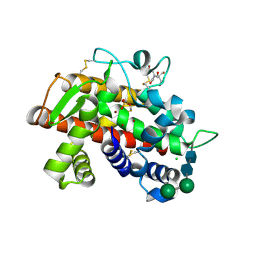 | | X-ray structure of fully glycosylated bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4CYE
 
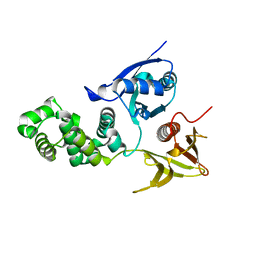 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZDT
 
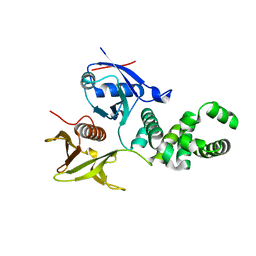 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3HUP
 
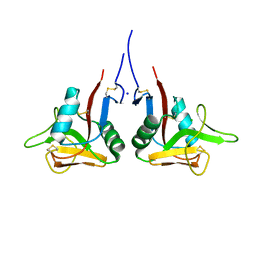 | | High-resolution structure of the extracellular domain of human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69, SODIUM ION | | Authors: | Kolenko, P, Dohnalek, J, Skalova, T, Hasek, J, Duskova, J, Vanek, O, Bezouska, K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | The high-resolution structure of the extracellular domain of human CD69 using a novel polymer
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3RS1
 
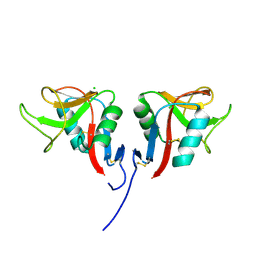 | | Mouse C-type lectin-related protein Clrg | | Descriptor: | C-type lectin domain family 2 member I, CHLORIDE ION | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
3RVA
 
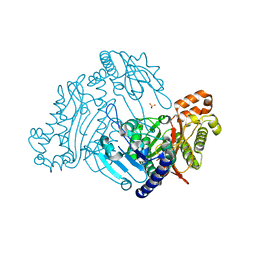 | | Crystal structure of organophosphorus acid anhydrolase from Alteromonas macleodii | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, Organophosphorus acid anhydrolase, ... | | Authors: | Stepankova, A, Koval, T, Ostergaard, L.H, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organophosphorus acid anhydrolase from Alteromonas macleodii: structural study and functional relationship to prolidases.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6AW6
 
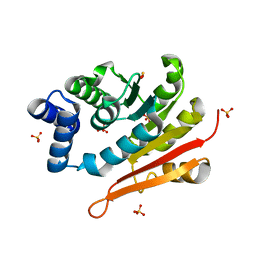 | | 1.70A resolution structure of catechol O-methyltransferase (COMT) L136M (rhombohedral form) from Nannospalax galili | | Descriptor: | Catechol O-methyltransferase, SULFATE ION | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW5
 
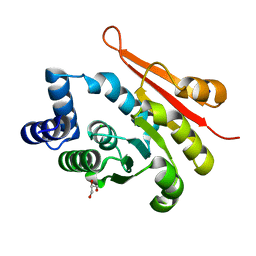 | | 1.90A resolution structure of catechol O-methyltransferase (COMT) L136M (hexagonal form) from Nannospalax galili | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW8
 
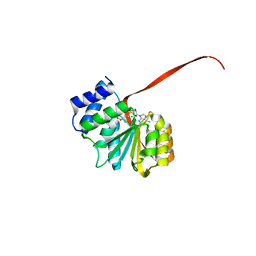 | | 2.25A resolution domain swapped dimer structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW4
 
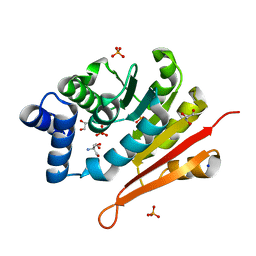 | | 1.50A resolution structure of catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW9
 
 | | 2.55A resolution structure of SAH bound catechol O-methyltransferase (COMT) L136M from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW7
 
 | | 2.15A resolution structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
