1PW5
 
 | | putative nagD protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, SULFATE ION, nagD protein, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-30 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | putative nagD protein
To be Published
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2G8Y
 
 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
8U7F
 
 | | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum | | Descriptor: | CIB_12 Beta-galactosidase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yakunin, A, Golyshin, P, Savchenko, A. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum
To Be Published
|
|
1SDJ
 
 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
2IGS
 
 | | Crystal Structure of the Protein of Unknown Function from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, GLYCEROL, Hypothetical protein, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-25 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Pseudomonas aeruginosa
To be Published
|
|
3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1PBJ
 
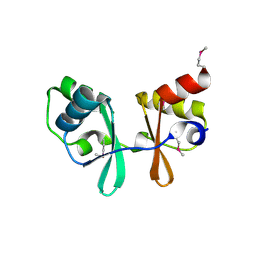 | | CBS domain protein | | Descriptor: | MAGNESIUM ION, hypothetical protein | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-14 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a hypothetical protein from M. thermautotrophicus reveals a
novel fold and a pseudo 2-fold axis of symmetry
TO BE PUBLISHED
|
|
3DR3
 
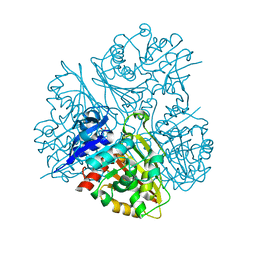 | | Structure of IDP00107, a potential N-acetyl-gamma-glutamylphosphate reductase from Shigella flexneri | | Descriptor: | D-MALATE, N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of IDP00107, a potential N-acetyl-gamma-glutamylphosphate reductase from Shigella flexneri
To be Published
|
|
3D3R
 
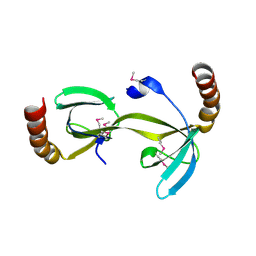 | | Crystal structure of the hydrogenase assembly chaperone HypC/HupF family protein from Shewanella oneidensis MR-1 | | Descriptor: | Hydrogenase assembly chaperone hypC/hupF | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hydrogenase Assembly Chaperone HypC/HupF Family Protein from Shewanella oneidensis MR-1.
To be Published
|
|
3DR6
 
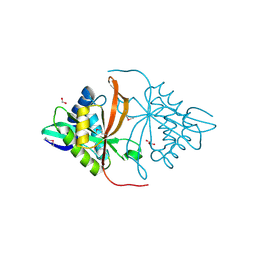 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
3FOV
 
 | | Crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris | | Descriptor: | NITRATE ION, UPF0102 protein RPA0323 | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-02 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris.
To be Published
|
|
2GFQ
 
 | | Structure of Protein of Unknown Function PH0006 from Pyrococcus horikoshii | | Descriptor: | MAGNESIUM ION, SULFATE ION, UPF0204 protein PH0006 | | Authors: | Cuff, M.E, Skarina, T, Gorodichtchenskaia, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of hypothetical protein ph0006 from Pyrococcus horikoshii
To be Published
|
|
3DR8
 
 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA
To be Published
|
|
3DRW
 
 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
3EXC
 
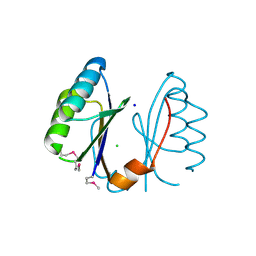 | | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Singer, A.U, Skarina, T, Tan, K, Kagan, O, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus
To be Published
|
|
3E8X
 
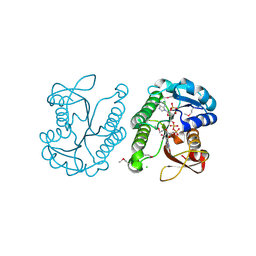 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
1SDI
 
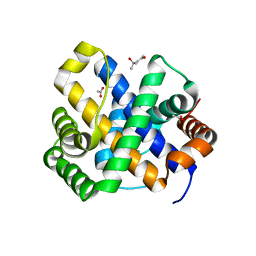 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
1SR8
 
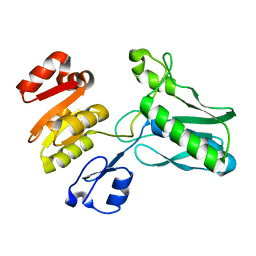 | | Structural Genomics, 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus | | Descriptor: | cobalamin biosynthesis protein (cbiD) | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus
To be Published
|
|
1MKZ
 
 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
