3GEU
 
 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
2O0Y
 
 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | Descriptor: | Transcriptional regulator | | Authors: | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
2ODK
 
 | | Putative prevent-host-death protein from Nitrosomonas europaea | | Descriptor: | GLYCEROL, Hypothetical protein, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crsytal structure of putative prevent-host-death protein from Nitrosomonas europaea.
To be Published
|
|
2NS0
 
 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
2OER
 
 | | Probable Transcriptional Regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Kim, Y, Skarina, T, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Probable Transcriptional Regulator from Pseudomonas aeruginosa
To be Published
|
|
1Y0N
 
 | | Structure of Protein of Unknown Function PA3463 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Hypothetical UPF0270 protein PA3463 | | Authors: | Binkowski, T.A, Edwards, A, Savchenko, A, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein PA3463 from Pseudomonas aeruginosa strain PAO1
To be Published
|
|
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2OR0
 
 | | Structural Genomics, the crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, Hydroxylase | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1
To be Published
|
|
2PJS
 
 | | Crystal structure of Atu1953, protein of unknown function | | Descriptor: | Uncharacterized protein Atu1953, ZINC ION | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily protein Atu1953, protein of unknown function
To be Published
|
|
2PYU
 
 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with IMP | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, Inosine Triphosphate Pyrophosphatase RdgB | | Authors: | Singer, A.U, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2FFS
 
 | | Structure of PR10-allergen-like protein PA1206 from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein PA1206 | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M.T, Wang, S, Kirillova, O, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PR10-Allergen-Like Protein PA1206 From Pseudomonas aeruginosa PAO1
To be Published
|
|
2FXA
 
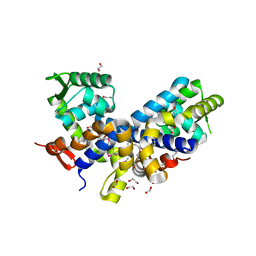 | | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis. | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis.
TO BE PUBLISHED
|
|
1WPB
 
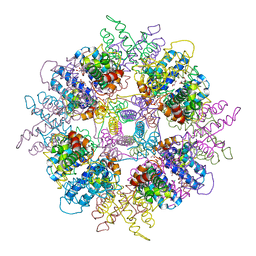 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
1S5U
 
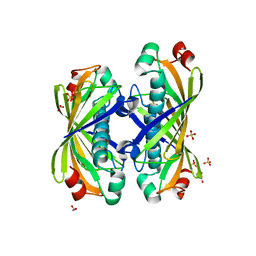 | | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Protein ybgC, SULFATE ION | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli
To be Published
|
|
5VIS
 
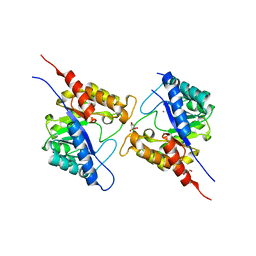 | | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium. | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Dihydropteroate Synthase, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil
Uncultured Bacterium.
To Be Published
|
|
2IA2
 
 | | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1 | | Descriptor: | Putative Transcriptional Regulator | | Authors: | Cymborowski, M.T, Chruszcz, M, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1
To be Published
|
|
5UXC
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXD
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5VRV
 
 | | 2.05 Angstrom Resolution Crystal Structure of C-terminal Domain (DUF2156) of Putative Lysylphosphatidylglycerol Synthetase from Agrobacterium fabrum. | | Descriptor: | GLYCEROL, Protein regulated by acid pH, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-11 | | Release date: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of C-terminal Domain (DUF2156) of Putative Lysylphosphatidylglycerol Synthetase from Agrobacterium fabrum.
To Be Published
|
|
5TPM
 
 | | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli. | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli.
To Be Published
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
1Z7A
 
 | | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1
To be Published
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
