1QL6
 
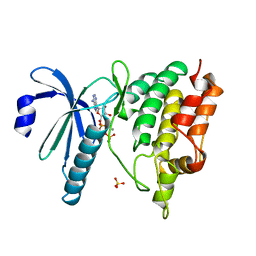 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
4G9M
 
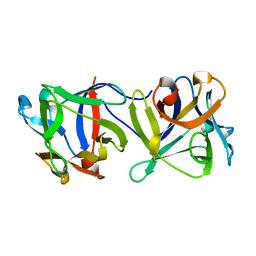 | |
4G9N
 
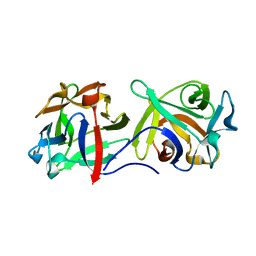 | |
3SYR
 
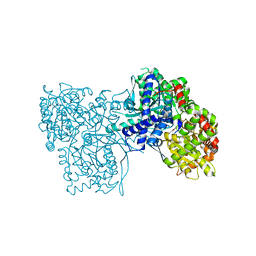 | | Glycogen phosphorylase b in complex with beta-D-glucopyranonucleoside 5-fluorouracil | | Descriptor: | 5-fluoro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Skamnaki, V.T, Kantsadi, A.L, Kontou, M, Leonidas, D.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3'-Axial CH(2) OH Substitution on Glucopyranose does not Increase Glycogen Phosphorylase Inhibitory Potency. QM/MM-PBSA Calculations Suggest Why.
Chem.Biol.Drug Des., 79, 2012
|
|
3SYM
 
 | | Glycogen Phosphorylase b in complex with 3 -C-(hydroxymethyl)-beta-D-glucopyranonucleoside of 5-fluorouracil | | Descriptor: | 5-fluoro-1-[3-C-(hydroxymethyl)-beta-D-glucopyranosyl]pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Skamnaki, V.T, Katsandi, A.L, Kontou, M, Leonidas, D.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3'-Axial CH(2) OH Substitution on Glucopyranose does not Increase Glycogen Phosphorylase Inhibitory Potency. QM/MM-PBSA Calculations Suggest Why.
Chem.Biol.Drug Des., 79, 2012
|
|
1W98
 
 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
3AMV
 
 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
3L7D
 
 | | Crystal Structure of Glycogen Phosphorylase DK5 complex | | Descriptor: | 1-(2,3-dideoxy-3-fluoro-beta-D-arabino-hexopyranosyl)-4-[(phenylcarbonyl)amino]pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3L7A
 
 | | Crystal Structure of Glycogen Phosphorylase DK2 complex | | Descriptor: | 1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)-4-[(phenylcarbonyl)amino]pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3L7B
 
 | | Crystal Structure of Glycogen Phosphorylase DK3 complex | | Descriptor: | 4-amino-1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3L79
 
 | | Crystal Structure of Glycogen Phosphorylase DK1 complex | | Descriptor: | 1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3L7C
 
 | | Crystal Structure of Glycogen Phosphorylase DK4 complex | | Descriptor: | 1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)-5-fluoropyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
2GPA
 
 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
2PHK
 
 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
2CCI
 
 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
8R53
 
 | | The complex of Glycogen Phosphorylase with (-)-Epigallocatechin-3-gallate (EGCG) and glucose. | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-15 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
8R52
 
 | | The complex of Glycogen Phosphorylase with epigallocatechin (EGC). | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-15 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
8R6V
 
 | | The complex of glycogen phosphorylase with EGCG (epigallocatechin gallate) and caffeine. | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-11-23 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
8QMU
 
 | | The complex of Glycogen Phosphorylase with (-)-Epigallocatechin-3-gallate (EGCG). | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Alexopoulos, S, Papakostopoulou, S, Koulas, M.S, Leonidas, D.D, Skamnaki, V. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Quercetin Binding Site of Glycogen Phosphorylase as a Target for Liver-Isoform-Selective Inhibitors against Glioblastoma: Investigation of Flavanols Epigallocatechin Gallate and Epigallocatechin.
J.Agric.Food Chem., 72, 2024
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
2JGZ
 
 | | Crystal structure of phospho-CDK2 in complex with Cyclin B | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN-B1 | | Authors: | Brown, N.R, Petri, E, Lowe, E.D, Skamnaki, V, Johnson, L.N. | | Deposit date: | 2007-02-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyclin B and cyclin A confer different substrate recognition properties on CDK2.
Cell Cycle, 6, 2007
|
|
1C50
 
 | | IDENTIFICATION AND STRUCTURAL CHARACTERIZATION OF A NOVEL ALLOSTERIC BINDING SITE OF GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Skamnaki, V.T, Tsitsanou, K.E, Gavalas, N.G, Johnson, L.N. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new allosteric site in glycogen phosphorylase b as a target for drug interactions.
Structure Fold.Des., 8, 2000
|
|
1E1Y
 
 | | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Flavopiridol Inhibits Glycogen Phosphorylase by Binding at the Inhibitor Site
J.Biol.Chem., 275, 2000
|
|
1C8K
 
 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
1C8L
 
 | | SYNERGISTIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG AND CAFFEINE | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, CAFFEINE, GLYCEROL, ... | | Authors: | Tsitsanou, K.E, Skamnaki, V.T, Oikonomakos, N.G. | | Deposit date: | 2000-05-16 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the synergistic inhibition of glycogen phosphorylase a by caffeine and a potential antidiabetic drug.
Arch.Biochem.Biophys., 384, 2000
|
|
