7S7J
 
 | | Structure of Human SPASTIN-IST1 complex. | | Descriptor: | CALCIUM ION, CHLORIDE ION, IST1 homolog, ... | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission.
Elife, 11, 2022
|
|
2LXL
 
 | | Lip5(mit)2 | | Descriptor: | Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
2LXM
 
 | | Lip5-chmp5 | | Descriptor: | Charged multivesicular body protein 5, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
4WZX
 
 | | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins | | Descriptor: | COBALT (II) ION, IST1 homolog, SULFATE ION, ... | | Authors: | Caballe, A, Wenzel, D.M, Agromayor, M, Alam, S.L, Skalicky, J.J, Kloc, M, Carlton, J.G, Labrador, L, Sundquist, W.I, Martin-Serrano, J. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3859 Å) | | Cite: | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Elife, 4, 2015
|
|
1YXR
 
 | | NMR Structure of VPS4A MIT Domain | | Descriptor: | vacuolar protein sorting factor 4A | | Authors: | Scott, J.A, Gaspar, J, Stuchell, M, Alam, S, Skalicky, J, Sundquist, W.I. | | Deposit date: | 2005-02-22 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of VPS4A MIT Domain
To be Published
|
|
5J9A
 
 | | Ambient temperature transition state structure of arginine kinase - crystal 11/Form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
5J99
 
 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
2K3W
 
 | | NMR structure of VPS4A-MIT-CHMP6 | | Descriptor: | Charged multivesicular body protein 6, Vacuolar protein sorting-associating protein 4A | | Authors: | Kieffer, C, Skalicky, J.J, Morita, E, De Domini, I, Ward, D.M, Kaplan, J, Sundquist, W.I. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding
Dev.Cell, 15, 2008
|
|
2JQH
 
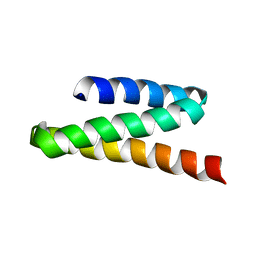 | | VPS4B MIT | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQ9
 
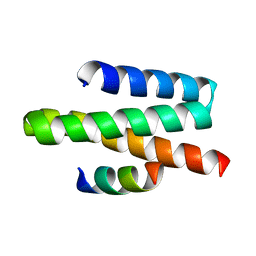 | | VPS4A MIT-CHMP1A complex | | Descriptor: | Chromatin-modifying protein 1a, Vacuolar protein sorting-associating protein 4A | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQK
 
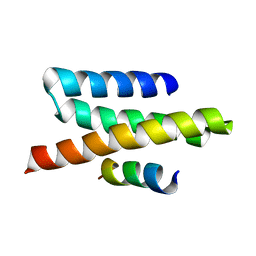 | | VPS4B MIT-CHMP2B Complex | | Descriptor: | Charged multivesicular body protein 2b, Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2KTZ
 
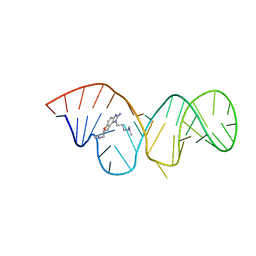 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2M7R
 
 | | Nmda receptor antagonist, conantokin bk-b, nmr, 20 structure | | Descriptor: | CONANTOKIN BK-B | | Authors: | Curtice, K.J, Platt, R.J, Skalicky, J.J, Horvath, M.P, Twede, V.D. | | Deposit date: | 2013-04-29 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes.
Toxicon, 81C, 2014
|
|
