2UYZ
 
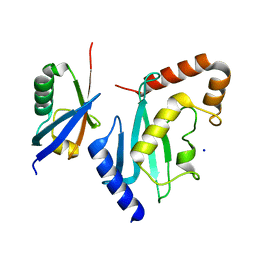 | | Non-covalent complex between Ubc9 and SUMO1 | | Descriptor: | SMALL UBIQUITIN-RELATED MODIFIER 1, SODIUM ION, SUMO-CONJUGATING ENZYME UBC9 | | Authors: | Knipscheer, P, van Dijk, W.J, Olsen, J.V, Mann, M, Sixma, T.K. | | Deposit date: | 2007-04-21 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation.
EMBO J., 26, 2007
|
|
2W8G
 
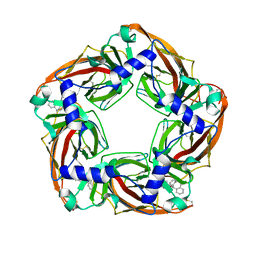 | | Aplysia californica AChBP bound to in silico compound 35 | | Descriptor: | (3-ENDO,8-ANTI)-8-BENZYL-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
1LTA
 
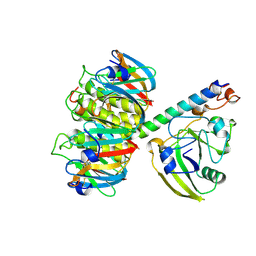 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | Authors: | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
4UEL
 
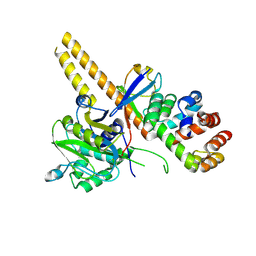 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | Descriptor: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UEM
 
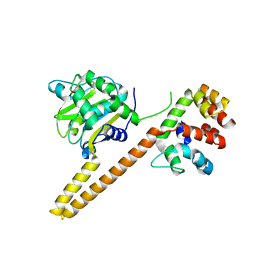 | | UCH-L5 in complex with the RPN13 DEUBAD domain | | Descriptor: | PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF5
 
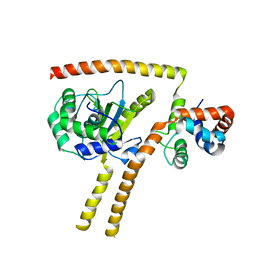 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF6
 
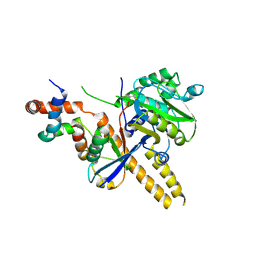 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
7AIC
 
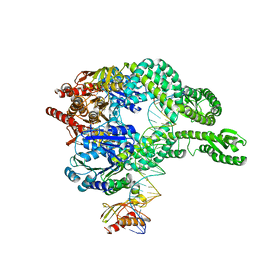 | | MutS-MutL in clamp state (kinked clamp domain) | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI7
 
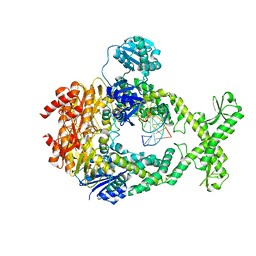 | | MutS in Intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIB
 
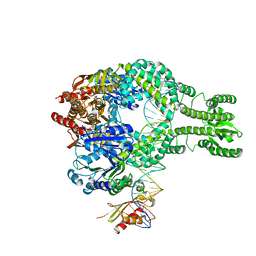 | | MutS-MutL in clamp state | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI6
 
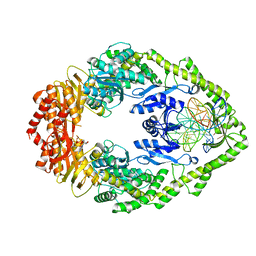 | | MutS in mismatch bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein MutS | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI5
 
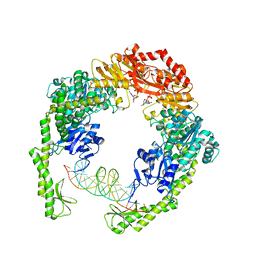 | | MutS in Scanning state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5AKB
 
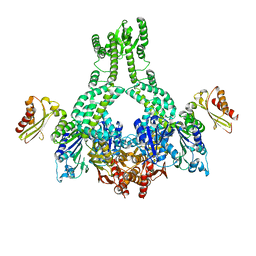 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKD
 
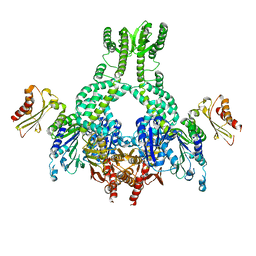 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKC
 
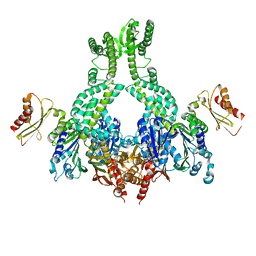 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
4AFT
 
 | | Aplysia californica AChBP in complex with Varenicline | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, VARENICLINE | | Authors: | Rucktooa, P, Haseler, C.A, vanElke, R, Smit, A.B, Gallagher, T, Sixma, T.K. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Characterization of Binding Mode of Smoking Cessation Drugs to Nicotinic Acetylcholine Receptors Through Study of Ligand Complexes with Acetylcholine-Binding Protein.
J.Biol.Chem., 287, 2012
|
|
4AEA
 
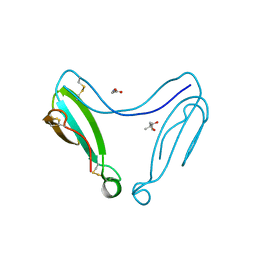 | | Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, LONG NEUROTOXIN 1 | | Authors: | Rucktooa, P, Osipov, A.V, Kasheverov, I.E, Filkin, S.Y, Starkov, V.G, Andreeva, T.V, Bertrand, D, Utkin, Y.N, Tsetlin, V.I, Sixma, T.K. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dimeric Alpha-Cobratoxin X-Ray Structure: Localization of Intermolecular Disulfides and Possible Mode of Binding to Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 287, 2012
|
|
4BQT
 
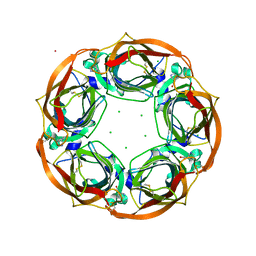 | | Aplysia californica AChBP in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Rucktooa, P, Haseler, C.A, vanElke, R, Smit, A.B, Gallagher, T, Sixma, T.K. | | Deposit date: | 2013-06-02 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Characterization of Binding Mode of Smoking Cessation Drugs to Nicotinic Acetylcholine Receptors Through Study of Ligand Complexes with Acetylcholine-Binding Protein.
J.Biol.Chem., 287, 2012
|
|
1LTB
 
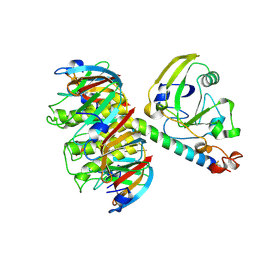 | |
4BFQ
 
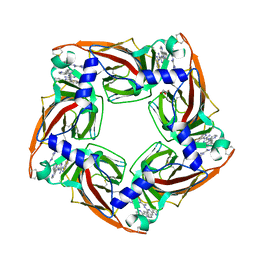 | | Assembly of a triple pi-stack of ligands in the binding site of Aplysia californica acetylcholine binding protein (AChBP) | | Descriptor: | 4,6-dimethyl-N'-(3-pyridin-2-ylisoquinolin-1-yl)pyrimidine-2-carboximidamide, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Stornaiuolo, M, De Kloe, G.E, Rucktooa, P, Fish, A, van Elk, R, Edink, E.S, Bertrand, D, Smit, A.B, de Esch, I.J.P, Sixma, T.K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly of a Pi-Pi Stack of Ligands in the Binding Site of an Acetylcholine Binding Protein
Nat.Commun., 4, 2013
|
|
1U9A
 
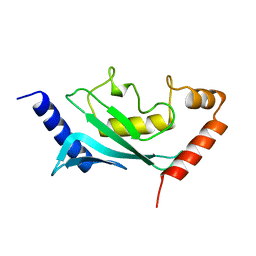 | | HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
4AYC
 
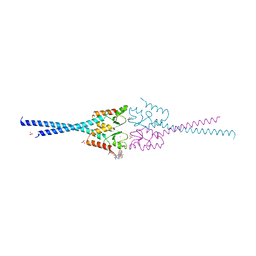 | | RNF8 RING domain structure | | Descriptor: | CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF8, GLYCEROL, ... | | Authors: | Mattiroli, F, Vissers, J.H.A, Van Dijk, W.J, Ikpa, P, Citterio, E, Vermeulen, W, Marteijn, J.A, Sixma, T.K. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-26 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rnf168 Ubiquitinates K13-15 on H2A/H2Ax to Drive DNA Damage Signaling
Cell(Cambridge,Mass.), 150, 2012
|
|
2C9T
 
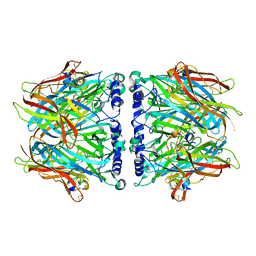 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1OH5
 
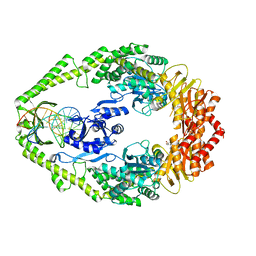 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
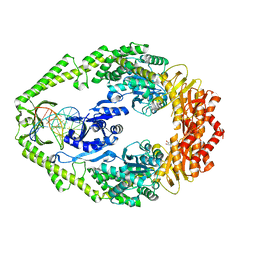 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
