3PB3
 
 | | Structure of an Antibiotic Related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
1KSL
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSV
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSK
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1MC3
 
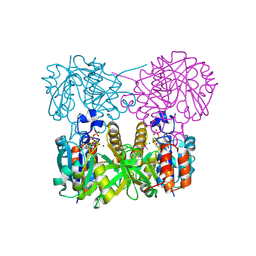 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
1EOJ
 
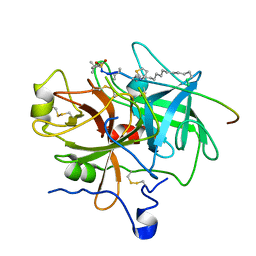 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
1EOL
 
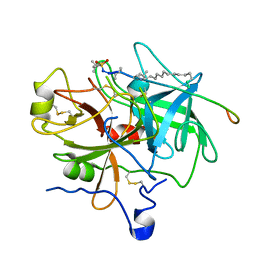 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P628 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
7Y6G
 
 | |
7Y6P
 
 | | Cryo-EM structure if bacterioferritin holoform | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
7Y6F
 
 | | Cryo-EM structure of Apo form of ScBfr | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
8H8X
 
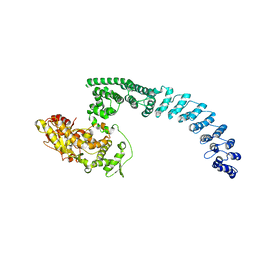 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8HAE
 
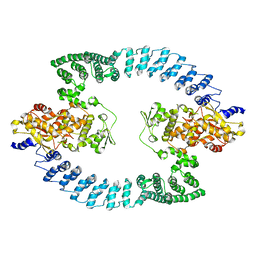 | | Cryo-EM structure of HACE1 dimer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8JAX
 
 | |
8JB0
 
 | |
4E53
 
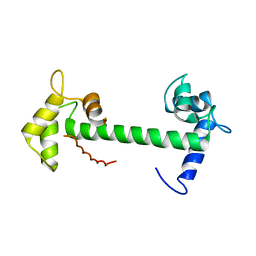 | | Calmodulin and Nm peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neuromodulin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
4E50
 
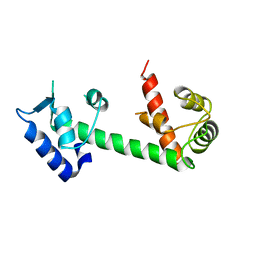 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
1CJL
 
 | |
8GWR
 
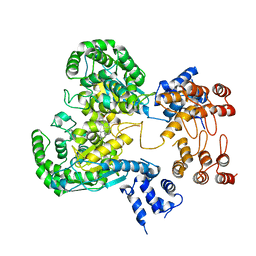 | | Near full length Kidney type Glutaminase in complex with 2,2-Dimethyl-2,3-Dihydrobenzo[a] Phenanthridin-4(1H)-one (DDP) | | Descriptor: | 2,2-dimethyl-1,3-dihydrobenzo[a]phenanthridin-4-one, Glutaminase kidney isoform, mitochondrial | | Authors: | Shankar, S, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A novel allosteric site employs a conserved inhibition mechanism in human kidney-type glutaminase.
Febs J., 290, 2023
|
|
8I54
 
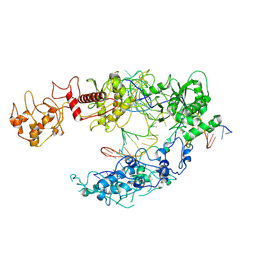 | | Lb2Cas12a RNA DNA complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3'), Lb2Cas12a, ... | | Authors: | Li, J, Sivaraman, J, Satoru, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
2GML
 
 | | Crystal Structure of Catalytic Domain of E.coli RluF | | Descriptor: | Ribosomal large subunit pseudouridine synthase F | | Authors: | Sunita, S, Zhenxing, H, Swaathi, J, Cygler, M, Matte, A, Sivaraman, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Organization and Crystal Structure of the Catalytic Domain of E.coli RluF, a Pseudouridine Synthase that Acts on 23S rRNA
J.Mol.Biol., 359, 2006
|
|
5JYP
 
 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | Descriptor: | 2-phenyl-~{N}-[5-[(1~{S},3~{S})-3-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]cyclohexyl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Ramachandran, S, Sivaraman, J. | | Deposit date: | 2016-05-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
7C28
 
 | | Unusual quaternary structure of a homodimeric synergistic toxin from mamba snake venom | | Descriptor: | SULFATE ION, Synergistic-type venom protein S2C4 | | Authors: | Jobichen, C, Narumi, A, Sivaraman, J, Kini, R.M. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual quaternary structure of a homodimeric synergistic-type toxin from mamba snake venom defines its molecular evolution.
Biochem.J., 477, 2020
|
|
2ED6
 
 | |
7CBK
 
 | | Structure of Human Neutrophil Elastase Ecotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.
Biochemistry, 59, 2020
|
|
3O4F
 
 | | Crystal Structure of Spermidine Synthase from E. coli | | Descriptor: | SULFATE ION, Spermidine synthase | | Authors: | Zhou, X, Tkaczuk, K.L, Chruszcz, M, Chua, T.K, Minor, W, Sivaraman, J. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Escherichia coli spermidine synthase SpeE reveals a unique substrate-binding pocket
J.Struct.Biol., 169, 2010
|
|
