1E2T
 
 | | Arylamine N-acetyltransferase (NAT) from Salmonella typhimurium | | Descriptor: | N-HYDROXYARYLAMINE O-ACETYLTRANSFERASE | | Authors: | Sinclair, J.C, Sandy, J, Delgoda, R, Sim, E, Noble, M.E.M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Nat.Struct.Biol., 7, 2000
|
|
4C5P
 
 | | The structure of mycobacterium marinum arylamine n-acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, AZIDE ION, GLYCEROL, ... | | Authors: | Abuhammad, A, Lowe, E.D, Garman, E.F, Sim, E. | | Deposit date: | 2013-09-13 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Arylamine N-Acetyltransferases from Mycobacteria: Investigations of a Potential Target for Anti- Tubercular Therapy
Ph D Thesis
|
|
1W6F
 
 | | Arylamine N-acetyltransferase from Mycobacterium smegmatis with the anti-tubercular drug isoniazid bound in the active site. | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Holton, S, Fullham, E, Sim, E, Noble, M.E.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Anti-Tubercular Drug Isoniazid to the Arylamine N-Acetyltransferase Protein from Mycobacterium Smegmatis
Protein Sci., 14, 2005
|
|
1W5R
 
 | | X-ray crystallographic structure of a C70Q Mycobacterium smegmatis N- arylamine Acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Holton, S.J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E.M, Sim, E. | | Deposit date: | 2004-08-09 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Investigation of the Catalytic Triad of Arylamine N-Acetyltransferases: Essential Residues Required for Acetyl Transfer to Arylamines.
Biochem.J., 390, 2005
|
|
1W4T
 
 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
8RKS
 
 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | Authors: | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | Deposit date: | 2023-12-30 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
8ABQ
 
 | | Structure of the SNX1-SNX5 complex, Pt derivative | | Descriptor: | PLATINUM (II) ION, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-07-04 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AFZ
 
 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8PYH
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Crinalium epipsammum PCC 9333 | | Descriptor: | ACETATE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZK
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8A1G
 
 | | Structure of the SNX1-SNX5 complex | | Descriptor: | N-PROPANOL, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7RS6
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to GMPCPP-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2024
|
|
7ZVR
 
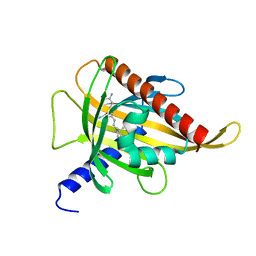 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZVQ
 
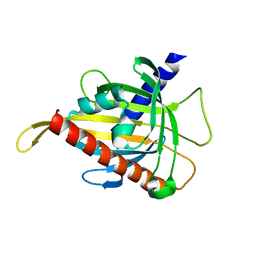 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7RS5
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to Taxol-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2021
|
|
6T6M
 
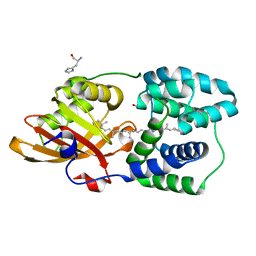 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T6K
 
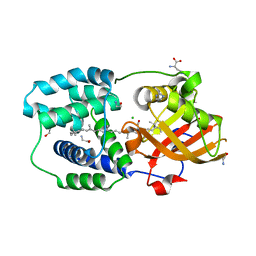 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
7ZXV
 
 | | Orange Carotenoid Protein Trp-288 BTA mutant | | Descriptor: | CHLORIDE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one, ... | | Authors: | Moldenhauer, M, Tseng, H.-W, Kraskov, A, Tavraz, N.N, Hildebrandt, P, Hochberg, G, Essen, L.-O, Budisa, N, Korf, L, Maksimov, E.G, Friedrich, T. | | Deposit date: | 2022-05-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parameterization of a single H-bond in Orange Carotenoid Protein by atomic mutation reveals principles of evolutionary design of complex chemical photosystems.
Front Mol Biosci, 10, 2023
|
|
8A0H
 
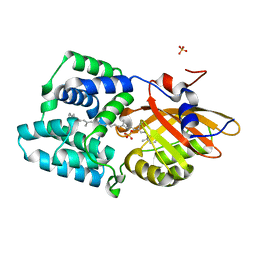 | | Crystal structure of the E25A mutant of the Orange Carotenoid Protein X from Gloeobacter kilaueensis JS1 complexed with echinenone | | Descriptor: | OCP N-terminal domain-containing protein, SULFATE ION, beta,beta-caroten-4-one | | Authors: | Boyko, K.M, Slonimskiy, Y.B, Zupnik, A.O, Varfolomeeva, L.A, Maksimov, E.G, Sluchanko, N.N. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A primordial Orange Carotenoid Protein: Structure, photoswitching activity and evolutionary aspects.
Int.J.Biol.Macromol., 222, 2022
|
|
7ZTU
 
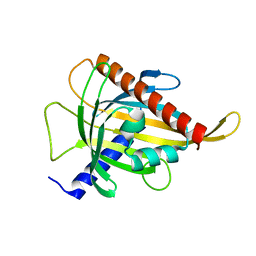 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, D162L mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZTQ
 
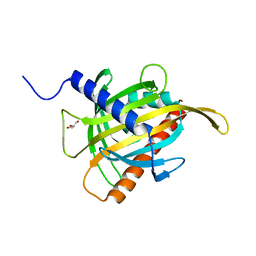 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform | | Descriptor: | Carotenoid-binding protein, GLYCEROL | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZTR
 
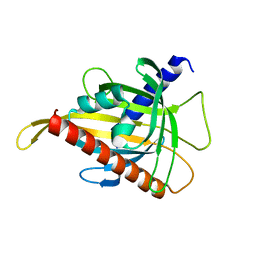 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
6T6O
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 4.6 | | Descriptor: | ASPARAGINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
4B1F
 
 | | Design of Inhibitors of Helicobacter pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty | | Descriptor: | 5-methyl-3-(1-methyl-1H-imidazol-5-yl)-7-(2-methylpropyl)-2-(naphthalen-1-ylmethyl)-2H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Basarab, G.S, Hill, P, Eyermann, C.J, Gowravaram, M, Kack, H, Osimoni, E. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Inhibitors of Helicobacter Pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles Onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3H8B
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
