8EX9
 
 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8CM3
 
 | |
4RCT
 
 | | Crystal structure of R-protein of NgoAVII restriction endonuclease | | Descriptor: | Restriction endonuclease R.NgoVII | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-17 | | Release date: | 2014-12-24 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4RD5
 
 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*GP*G)-3'), Restriction endonuclease R.NgoVII | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4RDM
 
 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3)', DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*G)-3)', Restriction endonuclease R.NgoVII, ... | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
6O9G
 
 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
8SUV
 
 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
6NSO
 
 | | An Unexpected Intermediate in the Reaction Catalyzed by Quinolinate Synthase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, IRON/SULFUR CLUSTER, Quinolinate synthase A | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
6NSU
 
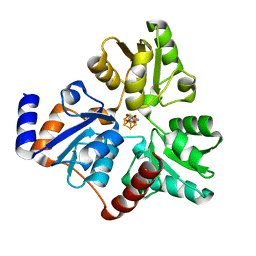 | | Crystallographic Capture of Quinolinate Synthase (NadA) from Pyrococcus horikoshii in its Substrates and Product-Bound States | | Descriptor: | DIDEHYDROASPARTATE, IRON/SULFUR CLUSTER, Quinolinate synthase A | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
6B77
 
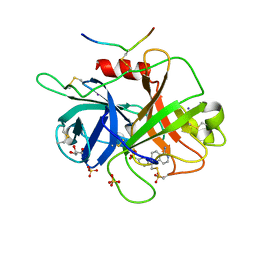 | | Structures of the two-chain human plasma factor XIIa co-crystallized with potent inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL, ... | | Authors: | Dementiev, A.A, Silva, A, Yee, C, Flavin, M.T, Partridge, J.R. | | Deposit date: | 2017-10-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors.
Blood Adv, 2, 2018
|
|
4QDL
 
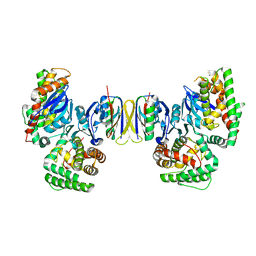 | | Crystal structure of E.coli Cas1-Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Tamulaitiene, G, Sinkunas, T, Silanskas, A, Gasiunas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2014-05-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli Cas1-Cas2 complex
To be Published
|
|
7BGS
 
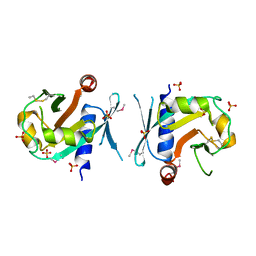 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PHO
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde | | Descriptor: | BICARBONATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Nunes-Costa, D, Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-17 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde
To Be Published
|
|
6ORA
 
 | | An Unexpected Intermediate in the Reaction Catalyzed by Quinolinate Synthase | | Descriptor: | (2~{Z})-2-(1-oxidanyl-3-oxidanylidene-propyl)iminobutanedioic acid, 2-hydroxy-N-[(1S)-1-hydroxy-3-oxopropyl]-L-aspartic acid, ACETATE ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
7OXF
 
 | |
8DV6
 
 | | Zika virus envelope protein structure in complex with a potent Human mAb | | Descriptor: | Envelope protein E, mAb Fab Heavy Chain, mAb Fab Light Chain | | Authors: | Cameron, A, Puhl, A.C, deSilva, A.M, Premkumar, L. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure and neutralization mechanism of a human antibody targeting a complex Epitope on Zika virus.
Plos Pathog., 19, 2023
|
|
4UIF
 
 | | Cryo-EM structure of Dengue virus serotype 2 in complex with antigen-binding fragments of human antibody 2D22 | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN PVP94 07 - ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.-N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
4UIH
 
 | | Cryo-EM structure of Dengue virus serotype 2 strain New Guinea-C complexed with human antibody 2D22 Fab at 37 degree C. The Fab molecules were added to the virus before 37 degree C incubation. | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN NEW GUINEA-C E PROTEIN ECTODOMAIN | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
1BO4
 
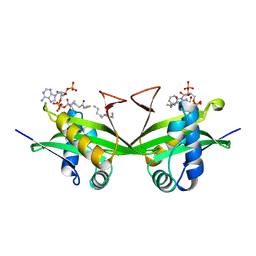 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
6THI
 
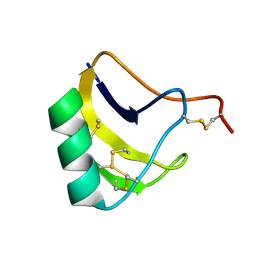 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
5MOU
 
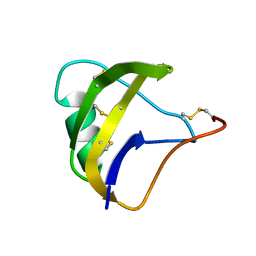 | | NMR spatial structure of scorpion alpha-like toxin BeM9 | | Descriptor: | Alpha-mammal toxin BeM9 | | Authors: | Mineev, K.S, Kuldushev, N.A, Berkut, A.A, Grishin, E.V, Vassilevski, A.A, Arseniev, A.S. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Proteins, 86, 2018
|
|
7QXJ
 
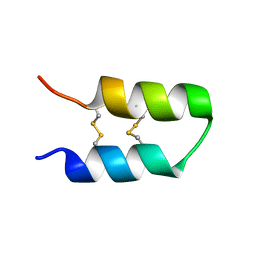 | |
7R0T
 
 | | Crystal structure of exonuclease ExnV1 | | Descriptor: | CHLORIDE ION, Exonuclease ExnV1, MAGNESIUM ION, ... | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Jasilionis, A, Linares-Pasten, J.A, Wang, L, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
